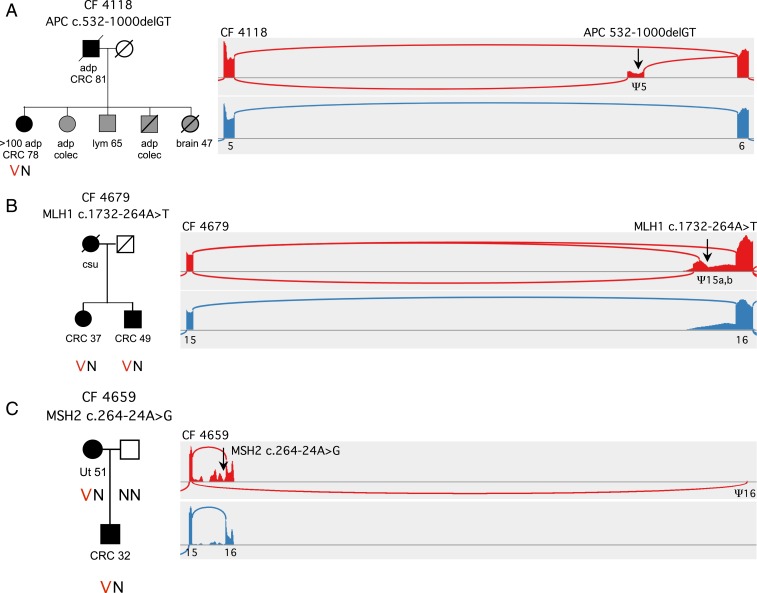
Fig. 3.
Analysis by cBROCA of intronic mutations leading to exonification. (A) In family CF4118, APC c.532-1000delGT (black arrow) leads to exonification of 165 bp in APC intron 5 and an in-frame stop. Exonification is very likely due to disruption of a silencing enhancer at chr5:112,115,485-112,115,492, and thereby activation of a splice acceptor at chr5:112,115,380 and of a splice donor at chr5:112,115,548. This exon does not appear in transcripts of controls. (B) In family CF4679, MLH1 c.1732-264A>T creates 2 exons that do not appear in normal MHL1 transcripts: one, with an insertion of 141 bp in the message and a stop at codon 587; and the other, with an insertion of 85 bp in the message and a stop at codon 581. The mechanism of exonification is likely creation of a donor splice at c.1732-264, activating previously silent acceptor sites at c.1732-406 and at c.1732-350. (C) In family CF4659, MSH2 c.2635-24A>G destroys a splicing branch-point, activating multiple cryptic splice sites in MSH1 intron 15, leading to multiple unstable transcripts. The mutation also activates a cryptic acceptor splice more than 30 kb downstream, at MSH2 c.2634+31432, creating transcripts that skip exon 16 and the 3′UTR, with a stop after 922 codons. Abbreviations are adenomatous polyps (adp), prophylactic colectomy (colec), colorectal cancer (CRC), lymphoma (lym), uterine cancer (Ut), cancer of unknown primary site (csu).