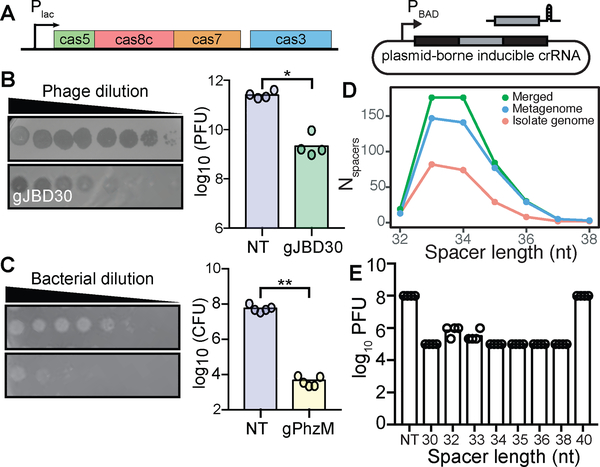
Figure 2. Heterologous expression in P. aeruginosa demonstrates the ability to target phage and chromosomal DNA.
(A) P. aeruginosa strain (PA01 tn7::lentaIC) constructed to inducibly express the minimal cas genes required for interference and a plasmid containing a minimal CRISPR array. (B) Expression of gJBD30 (phage-targeting) causes a 120-fold reduction in the number of plaque forming units (PFUs) when compared to a non-targeting (NT) control (n=4, *P=0.0286, Mann-Whitney U). (C) Expression of gPhzM (chromosome-targeting) decreases the colony forming units (CFUs) by 13,450-fold (n=5, **P=0.0079, Mann-Whitney U). (D) Distribution of spacer lengths found in the E. lenta isolate genomes, metagenomes, and merged datasets. (E) Variable length crRNAs decrease the number of PFUs with the exception of a 40 nt crRNA (n=4).