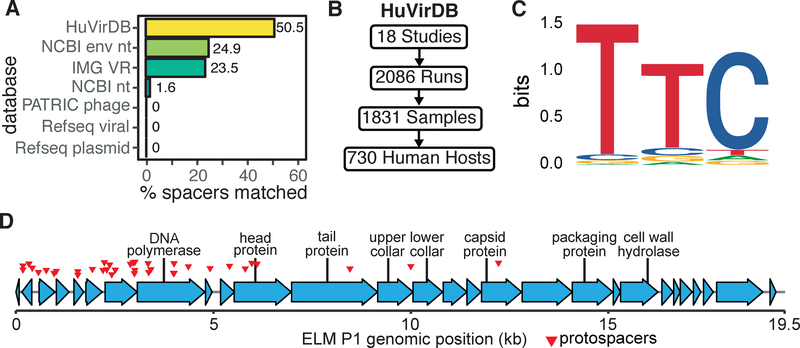
Figure 5. Discovering E. lenta predators based on protospacer enrichment.
(A) Comparison of protospacer matches within HuVirDB (249/493) versus other publicly accessible databases, including isolated and sequenced plasmids and phage from Refseq. (B) To facilitate phage discovery, public virome sequencing data was collected and assembled for our Human Virome Database (HuVirDB). (C) The 5’ flanking sequence was enriched for the canonical Type I-C protospacer adjacent motif (PAM) TTC. (D) Detailed annotation of a representative phage (ELM P1), identified based on a high frequency of matching E. lenta spacers.