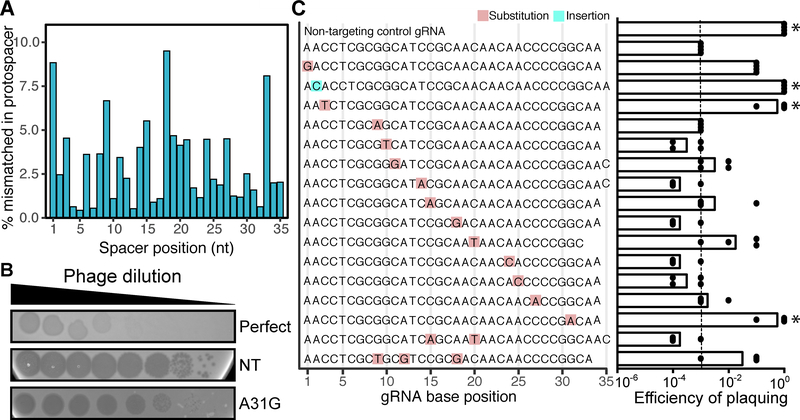
Figure 7. Most mismatches between the spacer and protospacer sequences still provide immunity against phage.
(A) The occurrence of mismatches throughout the length of spacer was calculated using HuVirDB. (B) Plaque assays revealed two phenotypes: opaque plaques (efficient targeting) and clearer plaques (poor targeting). Controls include: a perfect match positive control and a non-targeting (NT) negative control. A representative mismatch (A31G) is shown. (C) Plaquing efficiency [log-estimation in tested gRNA / log-estimation in NT control] reveals that mutations in the seed sequence of the crRNA allow the phage to escape CRISPR-Cas immunity (n = 4). The dashed line at 10−5 denotes the average value for the perfect match control gRNA. *P<0.05 Kruskal-Wallis with uncorrected Dunnett’s (compared to targeting control).