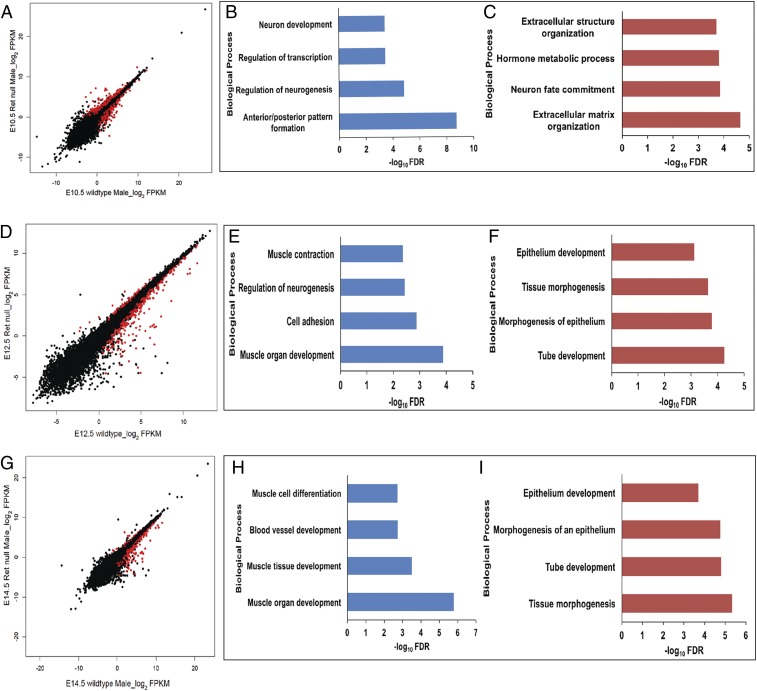
Fig. 6.
Ret-dependent gene expression in the developing gut. Scatterplots of log2 FPKM values between wild-type and Ret homozygous null embryonic gut at (A) E10.5, (D) E12.5, and (G) E14.5. Genes marked in red in the scatterplots have statistically significant (q value <0.01) expression differences between the indicated states. Genes activated by Ret at E10.5 (B) include neuronal genes and genes involved in transcriptional control, while those suppressed by Ret at E10.5 (C) include genes involved in extracellular matrix formation and in hormonal response. Similar annotation analyses at E12.5 show that along with neuronal genes, Ret also activates genes expressed in the musculature (E) and suppresses genes involved in epithelial development (F). At E14.5, Ret activates muscle-specific and vasculature development genes (H) while suppressing genes involved in epithelial morphogenesis (I).