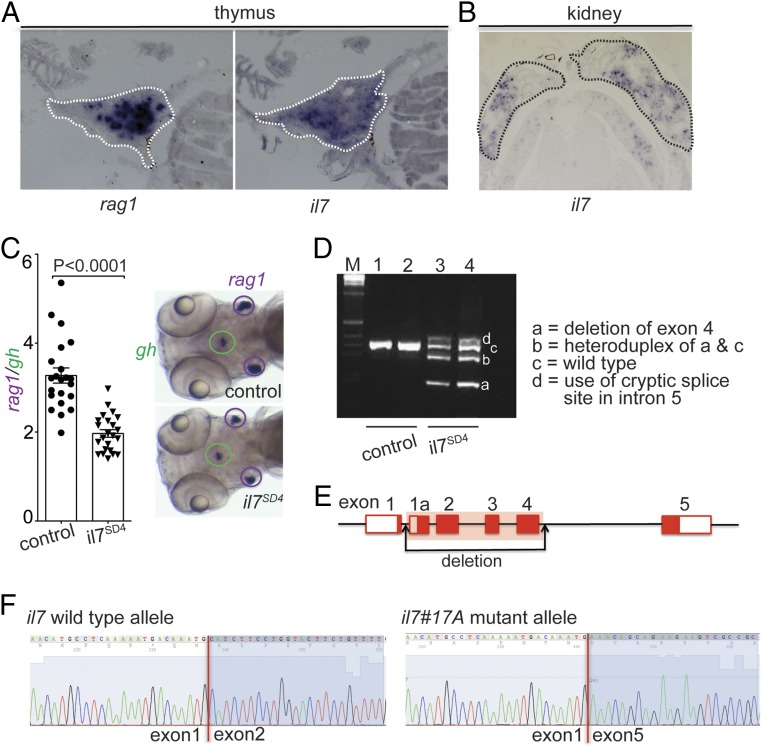
Fig. 1.
Characterization of the zebrafish il7 gene (ENSEMBL ID: ENSDARG00000092045). (A) RNA in situ hybridization on tissue sections of adult zebrafish thymus with probes specific for rag1 and il7. (B) RNA in situ hybridization on tissue sections of adult zebrafish kidney with a probe specific for il7. (C) Reduced thymopoietic activity in il7 morphants as determined by the thymopoietic index (Left), derived from quantitative analysis of whole-mount RNA in situ hybridization experiments (Right). Each data point represents 1 fish; data are mean ± SEM. (D) RT-PCR analysis of il7 cDNA structures resulting from interference with splicing of the pre-mRNA transcripts by an antisense oligonucleotide targeting the splice donor site of exon 4 (SD4). The deletion in variant a does not cause a frame shift, since all exons of the il7 gene are in phase 0; the use of cryptic splice site in the intron between exons 4 and 5 causes a frame shift mutation. (E) Schematic of the il7 gene structure, indicating the deletion introduced by CRISPR/Cas9 mutagenesis. (F) Sequence analysis of cDNAs demonstrating the loss of sequences derived from exons 2 to 4 in transcripts emanating from the il7#17A mutant allele.