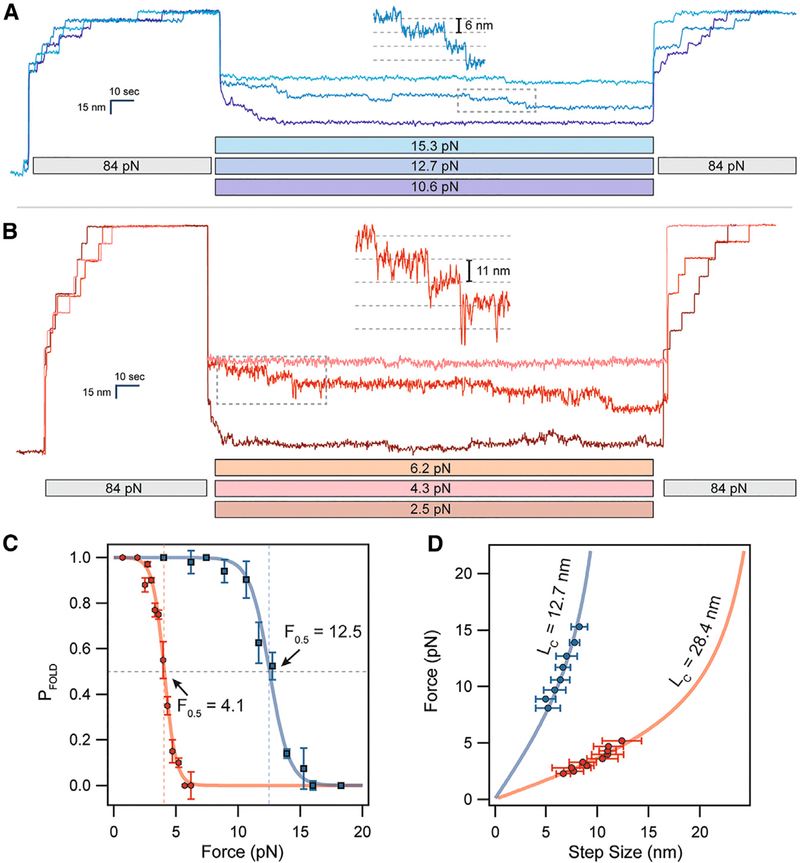
Figure 2. Cryptic Disulfide Bonds Alter the Folding Dynamics of Titin Ig Domains.
(A) (After full extension, oxidized polyproteins are allowed to fold by quenching the force to 10–16 pN for 100 s. While only a single folding event is observed at 16 pN, complete refolding occurs at 10.6 pN.
(B) Reduced polyproteins fold at a much lower range of forces in the 2–5 pN range.
(C) The folding probability is calculated as the number of refolded domains (probe pulse) divided by eight. Oxidation shifts the midpoint folding probability from 4.1 to 12.5 pN. The data were collected from several different molecules, with n = 11 for I27OXD and n = 9 for I27RED.
(D) The folding steps observed during the quench pulse (insets in A and B) scale in size with the applied force. Oxidized folding steps (blue circles) are fit by the FJC model (solid blue line, see also equations in STAR Methods) using a contour length of 12.7 nm. Reduced folding steps (red circles) are fit using a much larger contour length of 28.4 nm (solid red line). These data came from the same molecules measured in (C).