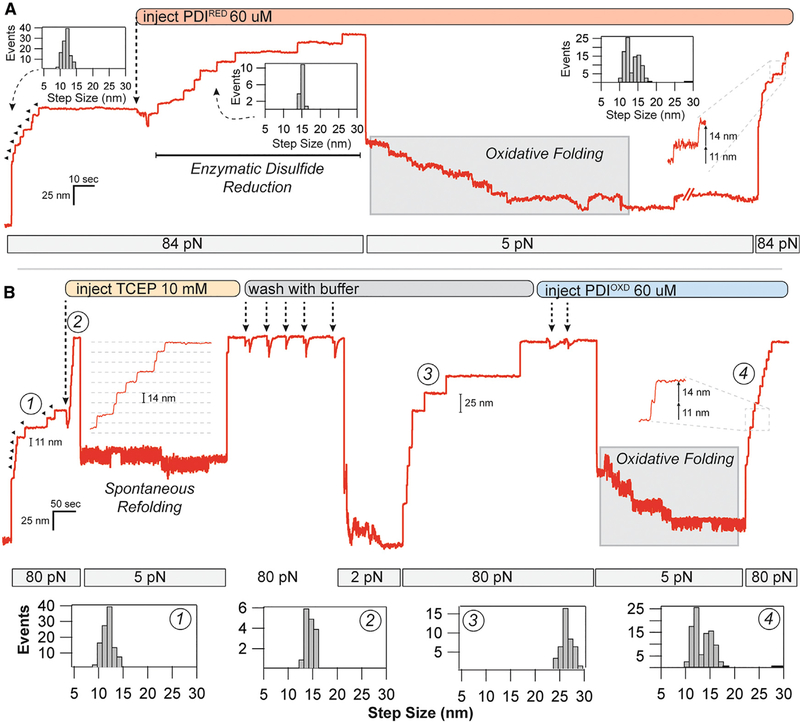
Figure 3. Oxidoreductase Mediated Disulfide Cleavage and Reformation under Force.
(A) After full unfolding of eight I27ox domains (11.4-nm steps), PDIRED was added to the flow cell, causing eight distinct disulfide cleavage events (14.6-nm steps). After quenching the force to 5 pN, a downward folding staircase is observed. Subsequent unfolding at 84 pN reveals a mixture of 11- and 14-nm steps, indicating reformation of the disulfide bond.
(B) Disulfide reformation could also be achieved with oxidized PDI. After full unfolding of eight I27OXD domains, TCEP (10 mM) was added to the flow cell, resulting in eight 14.6-nm steps due to the reduction of the disulfide bond. Quenching the force to 5 pN resulted in only a single refolding event. The protein was stretched again at high force to remove the TCEP with 53 washes using buffer. Reducing the force to 2 pN caused the refolding of 73 × I27RED domains, as indicated by the 25-nm steps. After full unfolding, PDIOXD was added to the flow cell, and the force was reduced to 5 pN again. In the presence of the oxidoreductase, a folding staircase is again observed. Subsequent pulling at 84 pN demonstrates 73 × 11- and 14-nm steps, indicating nearly complete reformation of the disulfide bond at 5 pN. Histograms in (A) and (B) are pooled from all of the oxidative folding experiments performed with the PDI oxidoreductase.