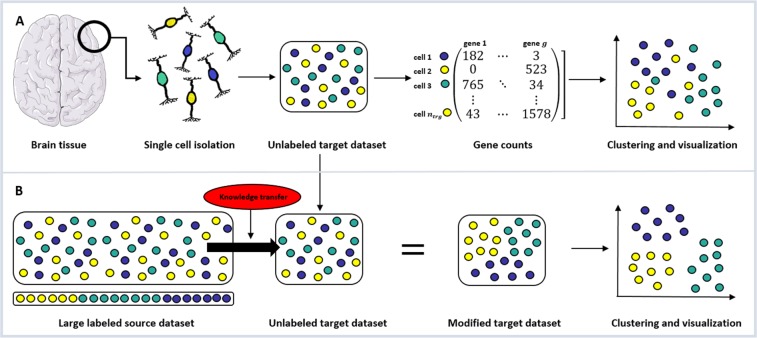
Figure 1.
Single-cell RNA sequencing (scRNA-Seq) and transfer learning. (A) Recent scientific and biotechnological developments have enabled scRNA-Seq, the accurate measurement of the transcriptional output of individual cells. Once a tissue sample (e.g. brain tissue) is extracted from an organism, single cells (e.g. neurons) are isolated and sequenced. For each gene the number of times a corresponding transcript is found in each individual cell is counted. These gene expression profiles of single cells are then used to identify tissue-specific cell types or states through an unsupervised clustering algorithm (e.g. SC3) which can eventually be visualized (through e.g. t-SNE or PCA plots). (B) When clustering smaller disease or tissue specific scRNA-Seq datasets it is often desirable to utilize large labeled reference datasets. The current work proposes to apply the machine learning concept of transfer learning to modify the unlabeled target dataset via Non-negative Matrix Factorization (NMF) in a way that reflects specific properties of a large labeled source dataset and improves the results of downstream clustering algorithms (in our case SC3). Please note, that even though this graph represents a complete overlap in cell types, both source and target datasets might include cell types that are not part of the other set. Graphs were created using Servier Medical Art (brain, neuron and syringe) according to a Creative Commons Attribution 3.0 Unported License guidelines 3.0 (https://creativecommons.org/licenses/by/3.0/). Colour changes were made to the original neuron cartoons.