Abstract
Background
Carbon (C) and nitrogen (N) are two fundamental components of starch and protein, which are important determinants of grain yield and quality. The food preferences of consumers and the expected end-use of grains in different rice-growing regions require diverse varieties that differ in terms of the grain N content (GNC) and grain C content (GCC) of milled rice. Thus, it is important that quantitative trait loci (QTLs)/genes with large effects on the variation of GNC and GCC are identified in breeding programs.
Results
To dissect the genetic basis of the variation of GNC and GCC in rice, the Dumas combustion method was used to analyze 751 diverse accessions regarding the GNC, GCC, and C/N ratio of the milled grains. The GCC and GNC differed significantly among the rice subgroups, especially between Xian/Indica (XI) and Geng/Japonica (GJ). Interestingly, in the GJ subgroup, the GNC was significantly lower in modern varieties (MV) than in landraces (LAN). In the XI subgroup, the GCC was significantly higher in MV than in LAN. One, six, and nine QTLs, with 55 suggestively associated single nucleotide polymorphisms, were detected for the GNC, GCC, and C/N ratio in three panels during a single-locus genome-wide association study (GWAS). Three of these QTLs were also identified in a multi-locus GWAS. We screened 113 candidate genes in the 16 QTLs in gene-based haplotype analyses. Among these candidate genes, LOC_Os01g06240 at qNC-1.1, LOC_Os05g33300 at qCC-5.1, LOC_Os01g04360 at qCN-1.1, and LOC_Os05g43880 at qCN-5.2 may partially explain the significant differences between the LAN and MV. These candidate genes should be cloned and may be useful for molecular breeding to rapidly improve the GNC, GCC, and C/N ratio of rice.
Conclusions
Our findings represent valuable information regarding the genetic basis of the GNC and GCC and may be relevant for enhancing the application of favorable haplotypes of candidate genes for the molecular breeding of new rice varieties with specific grain N and C contents.
Keywords: Grain nitrogen content, Grain carbon content, Germplasm, Genome-wide association study (GWAS), Rice (Oryza sativa L.), Haplotype analysis
Background
As a staple food for more than half of the global population, rice is one of the most widely grown cereals worldwide and is critical for food security. Additionally, it is the source of about 25–50% of the daily protein intake of humans in developing countries, especially in Asia (Deng et al. 2019). Carbon (C) and nitrogen (N) are two fundamental components of starch and protein, which are important determinants of grain yield and quality that influence the milling, appearance, eating and cooking qualities, nutritional qualities, and health benefits of grains (Martin and Fitzgerald 2002). The C/N ratio, which reflects the relative strength of C and N metabolism, is useful for evaluating the metabolic balance between C and N and the growth vigor in crop plants (Xu et al. 2018). Starch and protein account for 70–80 and 7–10% of the components in the rice endosperm, respectively. The synthesis of starch from sugars (or other carbohydrates) requires less energy than the production of other substances in the rice grain, and is conducive to dry-matter accumulation and high yield. The amino acid content in rice is relatively balanced compared with that of other crops, including wheat and maize (Peng et al. 2014). Thus, increasing the grain N content (GNC), grain C content (GCC), and C/N ratio in milled rice is very important for improving the rice nutritional quality and yield. The economic development in Asia, with China as an example, has altered the rice breeding strategy from blindly pursuing higher yield to paying equal attention to high yield and quality as well as decreasing production costs, but maintaining safety (Tang et al. 2017).
Amylose and amylopectin are two types of glucan polymers in starch that are synthesized via the synergistic effects of several enzymes (Jeon et al. 2010). Amylose is mainly composed of a linear chain of alpha-1,4-linked glucose residues, and is synthesized in a reaction catalyzed by the Wx-encoded granule-bound starch synthase I (Jeon et al. 2010). Genes at other loci, such as the du loci, that are under monogenic recessive control have an additive effect on lowering the amylose content (Yano et al. 1988). Amylopectin has a multiple-cluster structure comprising a highly branched glucan with alpha-1,6-glycosidic bonds, and its synthesis is coordinately catalyzed by the following three classes of enzymes: soluble starch synthases (SSs: SSI, SSIIa, and SSIIIa), starch branching enzymes (BEs: BEI, BEIIa, and BEIIb), and starch debranching enzymes (ISA1 and PUL) (Jeon et al. 2010). The OsSSI (SSS1) gene encodes starch synthase I, which affects the amylopectin structure, but has no significant effect on the amylose content (Kawakatsu et al. 2010b). The OsSSIIIa (Flo5) gene encodes starch synthase IIIa, which affects the amylopectin structure, amylose content, and physicochemical properties of rice grain starch (Zhou et al. 2016). Moreover, OsBEIIb encodes an amylase starch branching enzyme (SBE IIb) that influences the starch structure in rice endosperm (Lu and Park 2012; Yang et al. 2012).
Rice grain proteins can be categorized as functional proteins (approximately 10%) and seed storage proteins (SSPs; approximately 90%) (Yang et al. 2019). On the basis of solubility-linked physical properties, SSPs comprise the following four protein fractions: albumins, globulins, prolamins, and glutelins (Kawakatsu et al. 2008; Saito et al. 2012). Among these proteins, glutelins are the most abundant, accounting for about 60–80% of all SSPs (Makoto et al. 2003). Because of a higher lysine content and greater digestibility, the nutritional value of rice glutelin is superior to that of other rice storage proteins. Glutelins can be further divided into four groups (GluA, GluB, GluC, and GluD) based on their amino acid sequence similarity (Kawakatsu and Takaiwa 2010). Several glutelin genes have been cloned, including GluA, GluB1, GluB6, GluB7, GluD, and OsGZF1 (Kawakatsu et al. 2010a ; Wu et al. 2010; Yi et al. 2014). Additionally, considerable effort has been made toward dissecting the genetic mechanism underlying the rice grain protein content (GPC) (Ren et al. 2014; Terao and Hirose 2015; Tian et al. 2013; Wang et al. 2011; Wang et al. 2010a, b). However, the mechanism mediating the GPC differences remains relatively uncharacterized (Yang et al. 2019). The SSPs are controlled by complex multigene families (Tian et al. 2009). Interestingly, some of these genes not only regulate starch storage, but also affect the protein in the endosperm (She et al. 2010; Wang et al. 2011). The combined effects of OsAGPL2 and OsAGPS2b are very important for the accumulation of storage substances, such as starch and protein, in the rice endosperm (Tang et al. 2016).
Most of the above-mentioned isolated genes affecting the GPC and the grain starch content (GSC) were identified based on various rice mutants. Therefore, a few favorable alleles of these genes have been mined for rice breeding. Although many quantitative trait loci (QTLs) related to the GPC and GSC have been detected by linkage mapping (Cheng et al. 2013; Yao et al. 2017; Ye et al. 2010; Zhang et al. 2008; Zheng et al. 2011) and association studies (Chen et al. 2018; Wang et al. 2017; Xu et al. 2016), most of these studies focused on the crude protein and starch contents or the individual protein and starch fractions in milled rice. There have been no studies on the genetic mechanism controlling the GNC, GCC, and C/N ratio in milled rice.
In this study, a diverse panel consisting of 751 accessions from the 3000 Rice Genomes Project (3K RGP) (3K RGP 2014) was evaluated regarding the GNC and GCC in milled rice to identify related candidate genes in a genome-wide association study (GWAS) with high-density single nucleotide polymorphisms (SNPs). This was followed by a gene-based haplotype analysis. The objectives of our study were as follows: (1) screen representative resources with a distinct GNC, GCC, and C/N ratio in rice germplasm; (2) identify loci and candidate genes associated with the GNC, GCC, and C/N ratio; and (3) mine the favorable haplotypes/alleles of some important candidate genes in rice germplasm.
Results
Phenotypic Variations and Correlations
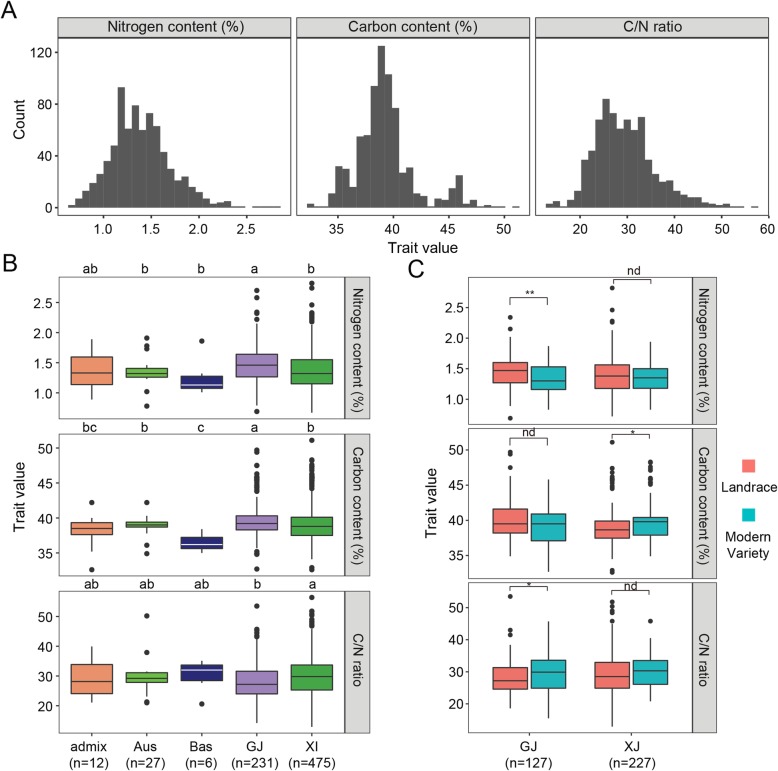
A diverse global collection of 751 Oryza sativa L. accessions were evaluated regarding their GNC, GCC, and calculated C/N ratios (Additional file 1: Table S1). A broad phenotypic distribution among the diverse rice accessions from the 3K RGP implied that substantial genetic variations controlled these three traits (Fig. 1a). The average GNC, GCC, and C/N ratio were 1.40% (0.67–2.82%), 39.3% (32.6–51.1%), and 29.5% (12.9–56.4%), respectively. An analysis of five rice subgroups revealed significant differences in these three traits, especially in the Xian/Indica (XI) and Geng/Japonica (GJ) comparisons (Fig. 1b). Specifically, the GNC (mean 1.47%; 0.69–2.70%) and GCC (mean 39.6%; 32.7–49.7%) of GJ were higher than the GNC (mean 1.36%; 0.67–2.82%) and GCC (mean 39.2%; 32.6–51.1%) of XI. In contrast, the C/N ratio of GJ (mean 28.0%; 14.2–53.5%) was lower than that of XI (mean 30.2%; 12.9–56.4%) (Fig. 1b and Additional file 1: Table S1). Additionally, the mean values of these three traits (GNC 1.35%, GCC 38.9%, and C/N ratio 29.6%) for Aus were similar to those of XI, which is consistent with their close phylogenetic relationship (Wang et al. 2018). However, significant differences in the GNC and GCC were observed between the basmati (Bas) and GJ subgroups, which also have a close phylogenetic relationship (Fig. 1b). Interestingly, in the GJ subgroup, the mean GNC (1.38%) of modern varieties (MV) was significantly lower than that (1.48%) of landraces (LAN), whereas there were no significant differences in the mean GNC between the XI MV and LAN (Fig. 1c). Regarding the GCC, in the XI subgroup, the mean value was significantly higher in the MV (39.9%) than in the LAN (39.1%). In contrast, there was no significant difference in the GCC of the GJ MV and LAN. These results suggest the GNC and GCC were affected by intense selective breeding for diverse targets in GJ and XI. A significant positive correlation between GNC and GCC was determined for the whole population (r = 0.30, P < 0.001), but the correlation was greater in XI (r = 0.33, P < 0.001) than in GJ (r = 0.22, P < 0.001), suggesting some differentiation in the GNC and GCC between the two rice subgroups (Additional file 2: Table S2). Moreover, significantly negative correlations between GNC and the C/N ratio and no correlations between GCC and the C/N ratio, respectively, were detected for the whole, XI, and GJ populations (Additional file 2: Table S2). These findings imply that the genetic basis of the GNC and GCC probably differs between the XI and GJ accessions.
Fig. 1.
Grain nitrogen content (GNC), grain carbon content (GCC), and the C/N ratio of milled rice and correlations among these traits in rice subgroups. a Phenotypic distribution of the GNC, GCC, and C/N ratio in the whole population. b The GNC, GCC, and C/N ratio in five rice subgroups. Different letters above the boxplots indicate significant differences among subgroups (P < 0.05) based on Duncan’s test. c Differences in the GNC, GCC, and C/N ratio between landraces and modern varieties in the GJ and XI subgroups. *, **, and nd indicate significant differences at P < 0.05, P < 0.01, and no significant difference, respectively (Student’s t-test)
Single-Locus GWAS for the GNC, GCC, and C/N Ratio
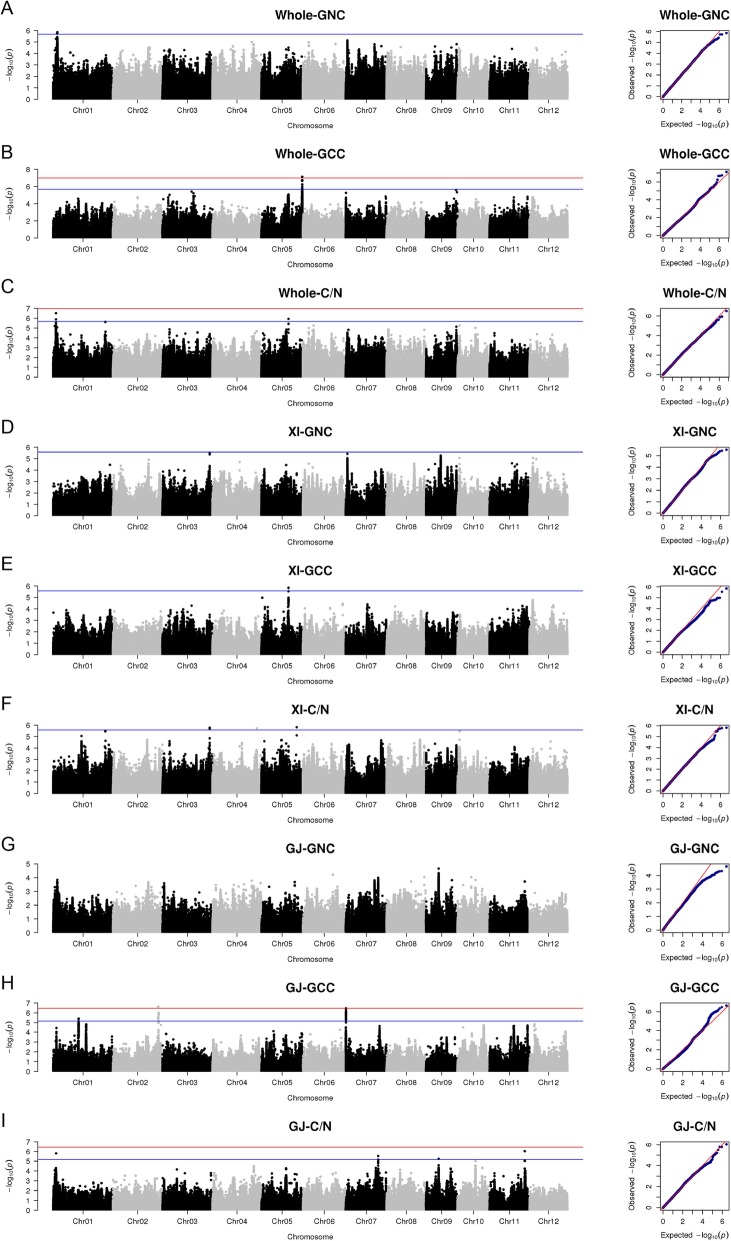
We conducted a single-locus GWAS to identify loci associated with the GNC, GCC, and C/N ratio in three panels (whole population, XI, and GJ) (Fig. 2). A total of 2,994,907, 2,118,326, and 1,318,493 filtered SNPs for the whole population, XI, and GJ panels, respectively, were used for the association analyses with the LMM of EMMAX (Kang et al. 2010). On the basis of a significant 1/N (N indicating the effective number of independent SNPs) calculated with the GEC software (Li et al. 2012), the Bonferroni-corrected genome-wide P-value thresholds of 2.09 × 10− 6, 2.66 × 10− 6, and 6.79 × 10− 6 were considered to reflect suggestive associations in the whole population, XI, and GJ panels, respectively (Additional file 3: Table S3). A total of 55 associated SNPs were detected on chromosomes 1, 2, 3, 4, 5, 7, 9, and 11 in all three panels, including 3, 40, and 12 SNPs associated with the GNC, GCC, and C/N ratio, respectively. These SNPs were located within or neighboring 38 annotated genes in the Nipponbare reference genome IRGSP 1.0 (Additional file 4: Table S4). We combined adjacent significantly associated SNPs within a linkage disequilibrium (LD) block as a QTL associated with the analyzed traits. Specifically, one (qNC-1.1), six (qCC-1.1, qCC-2.1, qCC-2.2, qCC-5.1, qCC-5.2, and qCC-7.1), and nine (qCN-1.1, qCN-1.2, qCN-3.1, qCN-4.1, qCN-5.1, qCN-5.2, qCN-7.1, qCN-9.1, and qCN-11.1) QTLs were detected for the GNC, GCC, and C/N ratio, respectively, in all panels (Table 1).
Fig. 2.
Manhattan and quantile-quantile plots for the single-locus GWAS. a Grain nitrogen content (NC), (b) grain carbon content (CC), and (c) the C/N ratio for the whole GWAS panel. d NC, (e) CC, and (f) the C/N ratio for the XI GWAS panel. g NC, (h) CC, and (i) the C/N ratio for the GJ GWAS panel. The points in the Manhattan plots indicate the −log10(P) value. The horizontal red and blue lines indicate the significant and suggestive thresholds calculated as follows: 0.05 and 1 divided by the effective number of independent markers in the GWAS panel, respectively
Table 1.
Sixteen QTLs for the GNC, GCC, and C/N ratio of milled rice identified in a single-locus GWAS
| Traits | QTL | Chromosome | Start (bp) | End (bp) | No. of significant SNP | Lead SNP | P value of lead SNP | Lead SNP located/flanking genes | Detected panelsa | Detected panels by multiple-locus GWAS | Known related genes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GNC | qNC-1.1 | 1 | 2,931,216 | 2,967,102 | 3 | rs1_2956867 | 1.38E-06 | LOC_Os01g06210-LOC_Os01g06220 | Whole | ||
| GCC | qCC-1.1 | 1 | 18,333,711 | 18,405,691 | 1 | rs1_18390016 | 3.95E-06 | LOC_Os01g33410 | GJ | ||
| qCC-2.1 | 2 | 32,773,074 | 32,956,207 | 3 | rs2_32881022 | 2.32E-07 | LOC_Os02g53710-LOC_Os02g53720 | GJ | |||
| qCC-2.2 | 2 | 33,160,172 | 33,274,475 | 4 | rs2_33,160,172 | 1.00E-06 | LOC_Os02g54120 | GJ | |||
| qCC-5.1 | 5 | 19,481,277 | 19,681,277 | 1 | rs5_19566944 | 1.44E-06 | LOC_Os05g33340 | XI | Whole | OsPPDKB | |
| qCC-5.2 | 5 | 29,438,881 | 29,580,663 | 8 | rs5_29562689 | 7.43E-08 | LOC_Os05g51550-LOC_Os05g51560 | Whole | NRR; CRCT | ||
| qCC-7.1 | 7 | 127,781 | 283,670 | 23 | rs7_144536 | 3.42E-07 | LOC_Os07g01240 | GJ | |||
| C/N ratio | qCN-1.1 | 1 | 1,942,481 | 1,952,710 | 2 | rs1_1952292 | 3.05E-07 | LOC_Os01g04380 | Whole | ||
| qCN-1.2 | 1 | 1,959,903 | 2,123,819 | 2 | rs1_2023817 | 1.57E-06 | LOC_Os01g04540 | GJ | |||
| qCN-3.1 | 3 | 34,300,375 | 34,309,240 | 2 | rs3_34300823 | 1.66E-06 | LOC_Os03g60310-LOC_Os03g60340 | XI | |||
| qCN-4.1 | 4 | 31,985,258 | 32,185,258 | 1 | rs4_32085258 | 1.93E-06 | LOC_Os04g53850 | XI | |||
| qCN-5.1 | 5 | 19,481,277 | 19,681,277 | 1 | rs5_19565967 | 1.14E-06 | LOC_Os05g33340 | Whole | OsPPDKB | ||
| qCN-5.2 | 5 | 25,421,042 | 25,621,042 | 1 | rs5_25521042 | 1.52E-06 | LOC_Os05g43880-LOC_Os05g43890 | XI | XI | ||
| qCN-7.1 | 7 | 23,515,915 | 23,515,983 | 1 | rs7_23515974 | 2.99E-06 | LOC_Os07g39280 | GJ | OsBZR1 | ||
| qCN-9.1 | 9 | 9,221,280 | 9,230,703 | 1 | rs9_9222530 | 5.93E-06 | LOC_Os09g15190-LOC_Os09g15200 | GJ | |||
| qCN-11.1 | 11 | 25,603,546 | 25,604,997 | 1 | rs11_25,603,546 | 9.05E-07 | LOC_Os11g42510-LOC_Os11g42520 | GJ | GJ |
aXI and GJ represent Xian/Indica and Geng/Japonica, respectively
Multi-Locus GWAS for the GNC, GCC, and C/N Ratio
In general, multiple testing correction methods, such as the Bonferroni correction method, for modifying the significant threshold value to control the false positive rate in a single-locus GWAS are so conservative that some associated SNPs may be eliminated. Therefore, we conducted a multi-locus association analysis with the mrMLM algorithm to solve this problem. This multi-locus GWAS detected more loci underlying the GNC, GCC, and C/N ratio than the single-locus GWAS for the same three panels. We identified 130 significant SNPs on all 12 chromosomes in at least one of the three panels, including 45, 34, and 51 SNPs for the GNC, GCC, and C/N ratio, respectively (Additional file 5: Table S5). For the GNC, 18, 27, and 4 SNPs were identified in the whole population, XI, and GJ panels, respectively, with the SNPs explaining 1.58–6.55%, 0.87–6.99%, and 7.00–14.79% of the phenotypic variations (PVE), respectively (Additional file 5: Table S5). Among these SNPs, four (rs4_1971938, rs4_30789977, rs6_23599588, and rs7_26720430) were detected in both the whole population and XI panels. Regarding the GCC, 11, 13, and 10 SNPs on all chromosomes, except for chromosome 10, were detected in the whole population, XI, and GJ panels, respectively, with PVE values of 1.34–5.78%, 2.43–5.20%, and 2.93–16.49%, respectively. For the C/N ratio, 18, 27, and 7 SNPs on all 12 chromosomes were detected in the whole population, XI, and GJ panels, respectively, with PVE values of 1.79–5.54%, 1.11–5.49%, and 4.40–8.16%, respectively. Among these associated SNPs, rs10_1541341 was simultaneously detected in the whole population and XI panels. Two QTLs, qCN-5.2 and qCN-11.1, detected in the single-locus GWAS were also identified as SNPs rs5_25521042 in the XI panel and rs11_25,603,546 in the GJ panel in the multi-locus GWAS. However, QTLs/genes related to the C/N ratio were not identified in these two regions, suggesting these regions may contain a potentially novel gene that should be finely mapped (Additional file 5: Table S5).
Haplotype Analyses for Candidate Genes
A total of 239 annotated genes located in the 16 QTLs detected in the single-locus GWAS underwent a haplotype analysis, and 113 genes were screened as candidate genes (Additional file 6: Table S6). These candidate genes were associated with at least eight plant metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Additional file 6: Table S6), including the flavonoid biosynthesis pathway. Flavonoids reportedly negatively affect starch synthesis in rice (Zhan et al. 2017). Four representative candidate genes were selected for the subsequent comprehensive analysis (Additional file 7: Table S7) according to the intensity of the association signals in the single-locus GWAS, the significance of the haplotype analyses (ANOVA), the biochemically related functions, and the expression profiles.
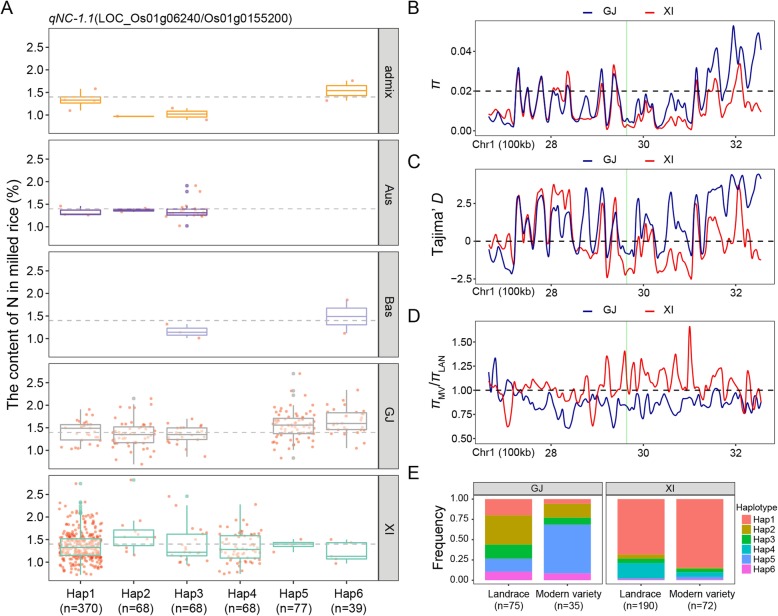
For the GNC, six genes annotated based on the Nipponbare reference genome IRGSP 1.0 with at least two haplotypes at qNC-1.1 (position 2,931,216 to 2,967,102 bp on chromosome 1) were concatenated by SNPs within the gene coding sequence region. Specifically, LOC_Os01g06240, which encodes a protein kinase, was detected as the candidate gene with the most significant differences (P = 9.80E-08) in the mean GNC among six haplotypes (Additional file 6: Table S6). The frequencies of all six haplotypes were significantly associated with the rice subgroups according to Fisher’s exact tests (Additional file 7: Table S7). Additionally, 94.8 and 76.9% of the accessions with the high-GNC haplotypes Hap5 (n = 77) and Hap6 (n = 39), respectively, as well as 77.9% of the accessions with the low-GNC haplotype Hap2 (n = 68) belonged to the GJ subgroup. In contrast, 90.3 and 100% of the accessions with the low-GNC haplotypes Hap1 (n = 370) and Hap4 (n = 68), respectively, belonged to the XI subgroup (Fig. 3a and Additional file 7: Table S7). Moreover, in the GJ subgroup, the frequency of Hap5 increased from 0.16 in LAN to 0.60 in MV, whereas the frequencies of the other four haplotypes (Hap1, Hap2, Hap3, and Hap6) were lower in MV than in LAN (Fig. 3). In the XI subgroup, the frequency of Hap1 increased slightly from 0.68 in LAN to 0.85 in MV. We analyzed the nucleotide diversity (π) and Tajima’s D statistics for a 600-kb region flanking LOC_Os01g06240 in the XI and GJ subgroups (Fig. 3b, c). The πGJ and πXI were similarly lower for the LOC_Os01g06240 region than for the flanking region, ranging from 0.002 to 0.005 (Fig. 3b). In terms of the allele frequency distributions, Tajima’s D was significantly more negative at LOC_Os01g06240 in the XI subgroup than in the GJ subgroup (Fig. 3c), implying there is an excess of rare alleles in the XI subgroup. Moreover, the πMV/πLAN ratio for LOC_Os01g06240 was 1.226 and 0.838 in the XI and GJ subgroups, respectively (Fig. 3d), suggesting LOC_Os01g06240 may have been affected by selective breeding more in the GJ subgroup than in the XI subgroup.
Fig. 3.
Haplotype analyses for the GNC and nucleotide diversity of the LOC_Os01g06240 candidate gene at qNC-1.1. a Haplotype analyses and comparisons of the mean GNC vs LOC_Os01g06240 haplotypes in five rice subgroups. Each red point around each boxplot indicates the GNC of one accession with the relative haplotype in its subgroup. Haplotypes in fewer than 10 accessions are not shown. b Nucleotide diversity (π), (c) Tajima’s D statistics, and (d) the ratio of the nucleotide diversity between modern varieties and landraces for the 600-kb genomic region flanking LOC_Os01g06240. Red and blue lines represent XI and GJ subgroups, respectively. The translucent green rectangle represents the LOC_Os01g06240 genomic region. e Frequencies of different haplotypes of LOC_Os01g06240 in XI and GJ landraces and modern varieties
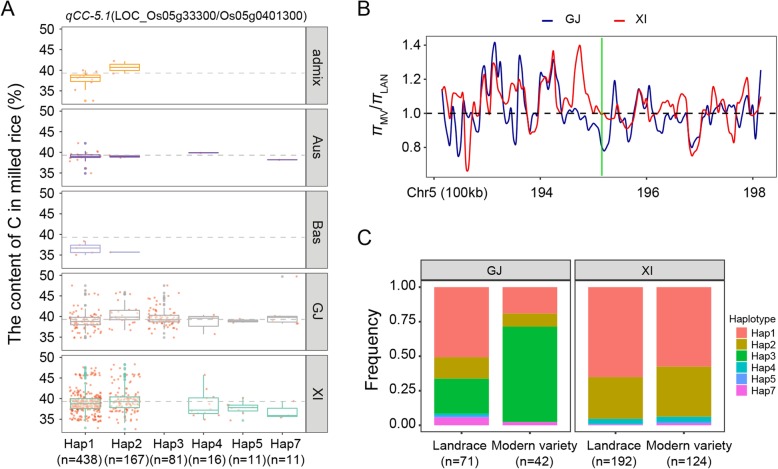
Regarding the GCC, our haplotype analysis revealed four, three, eight, two, and three candidate genes at the qCC-2.1, qCC-2.2, qCC-5.1, qCC-5.2, and qCC-7.1 QTLs, respectively (Additional file 6: Table S6). Additionally, LOC_Os05g33300, which encodes a Tat pathway signal sequence family protein, was detected as the candidate gene with the most significant differences (P = 4.90E-03) in the mean GCC among six haplotypes carried by at least 10 accessions (Additional file 6: Table S6). Moreover, 81.4 and 100% of the accessions with the representative high-GCC haplotypes Hap2 (n = 167) and Hap3 (n = 81) belonged to the XI and GJ subgroups, respectively. The frequency distributions of these two haplotypes differed significantly between the XI and GJ subgroups (Fig. 4a and Additional file 7: Table S7). Furthermore, the πMV/πLAN ratio for LOC_Os05g33300 was 0.83 and 1.00 in the GJ and XI subgroups, respectively (Fig. 4b). In the XI subgroup, the frequency of Hap2 increased from 0.30 in LAN to 0.36 in MV, whereas the frequency of Hap1 (low-GCC haplotype) decreased from 0.65 in LAN to 0.58 in MV (Fig. 4c). These results likely partially explain the greater GCC in MV than in LAN in the XI subgroup (Fig. 1c).
Fig. 4.
Haplotype analyses for the GCC and nucleotide diversity of the LOC_Os05g33300 candidate gene at qCC-5.1. a Haplotype analyses and comparisons of the mean GCC vs LOC_Os05g33300 haplotypes in five rice subgroups. Each red point around each boxplot indicates the GCC of one accession with the relative haplotype in its subgroup. Haplotypes in fewer than 10 accessions are not shown. b Ratio of the nucleotide diversity between modern varieties and landraces for the 600-kb genomic region flanking LOC_Os05g33300. Red and blue lines represent XI and GJ subgroups, respectively. The translucent green rectangle represents the LOC_Os05g33300 genomic region. c Frequencies of different haplotypes of LOC_Os05g33300 in XI and GJ landraces and modern varieties
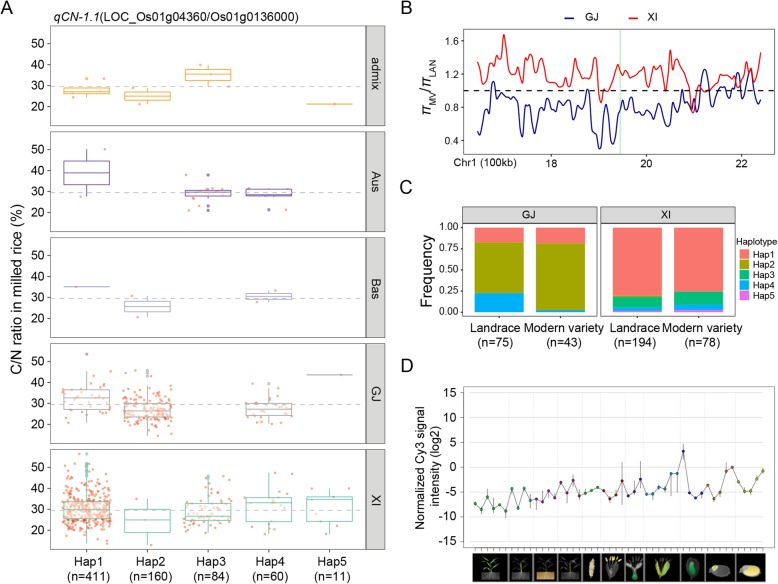
For the C/N ratio, 2, 35, 2, 19, 11, 20, 1, and 1 candidate genes were detected at the qCN-1.1, qCN-1.2, qCN-3.1, qCN-4.1, qCN-5.1, qCN-5.2, qCN-9.1, and qCN-11.1 QTLs, respectively, based on our haplotype analysis (Additional file 6: Table S6). Additionally, LOC_Os01g04360 at qCN-1.1 and LOC_Os05g43880 at qCN-5.2 were screened as important candidate genes with significant differences (P = 6.20E-08 and P = 6.90E-10) in the mean C/N ratio among different haplotypes in at least 10 accessions (Figs. 5 and 6 and Additional file 6: Table S6). The LOC_Os01g04360 candidate gene, which encodes a hsp20/alpha crystallin family protein, is highly expressed in specific organs (ovary, embryo, and endosperm) according to a publicly available rice gene expression profile database [RiceXPro (version 3.0)] (Fig. 5d). A comparison of the C/N ratios for the five haplotypes revealed that the representative high-C/N and low-C/N haplotypes were Hap1 and Hap2, with mean C/N ratios of 30.6 and 26.9%, respectively (Additional file 7: Table S7). Moreover, Hap1 and Hap2 were the major haplotypes in the XI and GJ subgroups, respectively, with significantly different frequency distributions between the two subgroups (Additional file 7: Table S7). We determined that 368 of 411 accessions (89.5%) with Hap1 belonged to the XI subgroup, whereas 153 of 160 accessions (95.6%) with Hap2 belonged to the GJ subgroup. The πMV/πLAN ratio for LOC_Os01g04360 was 0.73 and 1.27 in the GJ and XI subgroups, respectively (Fig. 5b). Furthermore, in the GJ subgroup, the frequency of Hap2 increased from 0.60 in LAN to 0.79 in MV, whereas the frequency of Hap4 (relatively low-C/N haplotype) decreased from 0.23 in LAN to 0.02 in MV (Fig. 5c).
Fig. 5.
Haplotype analyses for the C/N ratio and nucleotide diversity of the LOC_Os01g04360 candidate gene at qCN-1.1. a Haplotype analyses and comparisons of the mean C/N ratio vs LOC_Os01g04360 haplotypes in five rice subgroups. Each red point around each boxplot indicates the C/N ratio of one accession with the relative haplotype in its subgroup. Haplotypes in fewer than 10 accessions are not shown. b Ratio of the nucleotide diversity between modern varieties and landraces for the 600-kb genomic region flanking LOC_Os01g04360. Red and blue lines represent XI and GJ subgroups, respectively. The translucent green rectangle represents the LOC_Os01g04360 genomic region. c Frequencies of different haplotypes of LOC_Os01g04360 in XI and GJ landraces and modern varieties. d Normalized spatio-temporal expression of LOC_Os01g04360 in various Nipponbare tissues throughout the entire growth period in the field [downloaded from RiceXPro (version 3.0)]
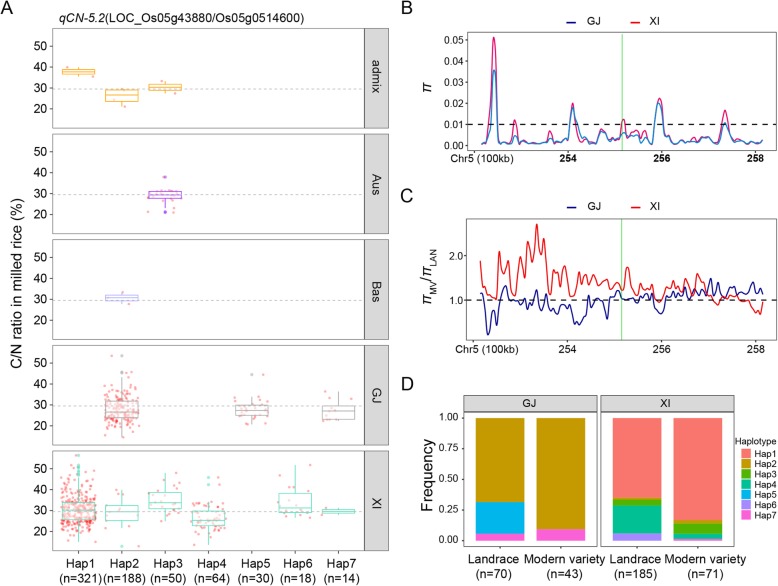
Fig. 6.
Haplotype analyses for the C/N ratio and nucleotide diversity of the LOC_Os05g43880 candidate gene at qCN-5.2. a Haplotype analyses and comparisons of the mean C/N ratio vs LOC_Os05g43880 haplotypes in five rice subgroups. Each red point around each boxplot indicates the C/N ratio of one accession with the relative haplotype in its subgroup. Haplotypes in fewer than 10 accessions are not shown. b Nucleotide diversity (π) and (c) the ratio of the nucleotide diversity between modern varieties and landraces for the 600-kb genomic region flanking LOC_Os05g43880. Red and blue lines represent XI and GJ subgroups, respectively. The translucent green rectangle represents the LOC_Os05g43880 genomic region. d Frequencies of different haplotypes of LOC_Os05g43880 in XI and GJ landraces and modern varieties
The LOC_Os05g43880 sequence (encoding a gibberellin 2-beta-dioxygenase) was slightly more diverse in the XI subgroup than in the GJ subgroup (Fig. 6b). The πMV/πLAN ratio for LOC_Os05g43880 in the XI subgroup was a little higher than that in the GJ subgroup (Fig. 6c). Multiple comparisons of the C/N ratios for the seven haplotypes indicated the representative high-C/N and low-C/N haplotypes were Hap6 and Hap4, with mean C/N ratios of 34.1 and 26.5%, respectively (Additional file 6: Table S6). The relatively low-C/N haplotypes (Hap2, Hap5, and Hap7) were significantly more abundant in the GJ subgroup, whereas the relatively high-C/N haplotypes (Hap1, Hap3, and Hap6) as well as one relatively low-C/N haplotype (Hap4) were mainly detected in the XI subgroup (Fig. 6a and Additional file 7: Table S7). These results partially explain the significant differences in the C/N ratio between XI and GJ (Fig. 1b). Furthermore, in the GJ subgroup, the frequency of Hap2 (mean C/N ratio of 28.0%) increased from 0.69 in LAN to 0.91 in MV, whereas the frequency of Hap5 (mean C/N ratio of 27.8%) decreased from 0.23 in LAN to 0 in MV (Fig. 6d), which partially explains the significant differences in the C/N ratio between LAN and MV in this subgroup (Fig. 1c).
Discussion
Simultaneous High-Throughput Phenotyping for GNC and GCC
The high-throughput and accurate phenotyping for target traits is currently more important and challenging than the genotyping by next-generation sequencing in a large-scale GWAS. In this study, we used an elemental analyzer based on the Dumas combustion method to rapidly and accurately determine the GNC and GCC in a large population set comprising 751 rice accessions. The application of the Dumas combustion method for analyzing cereals reportedly produces satisfactorily accurate results over a long period (Beljkaš et al. 2010). The repeatability and reproducibility standard deviations for analyses of cereals are lower than required by the Association of Official Analytical Chemists (Beljkaš et al. 2010). Compared with the Kjeldahl method (Beljkaš et al. 2010) for analyzing GPC and the chemical oxidation method (Isabella et al. 2004) for analyzing GCC, the Dumas combustion method is simpler, faster, and produces fewer system errors when simultaneously analyzing the GNC, GCC, and C/N ratio of one sample. Thus, despite the considerable cost and the equipment required for the Dumas combustion method, it is suitable for the high-throughput phenotyping for GNC and GCC in milled rice.
The GNC and GCC Were Affected by Diverse Selective Breeding in XI and GJ
Grain quality, which is a complex trait controlled by multiple genes, influences the milling, appearance, eating and cooking qualities, and nutritional qualities of rice. The nutritional quality of rice is mainly affected by the GPC and amino acid composition. Protein is the second most abundant component of rice grains, accounting for 7–10% of the rice endosperm dry weight (Martin and Fitzgerald 2002). The GPC is generally believed to be negatively correlated with the palatability and cooking quality of rice (Ning et al. 2010). A high GPC may lead to densely structured rice grains, which will result in hard and loose cooked rice (i.e., poor palatability) (Martin and Fitzgerald 2002). However, a high GPC will result in rice grains with a high nutritional quality (Long et al. 2013). Thus, the breeding targets for improving rice grain quality largely depend on the food preferences of consumers and the expected end-use of grains in various rice-growing regions worldwide. In this study, the GCC and GNC, which are two fundamental starch and protein characteristics that usually affect rice grain quality, were significantly different among the analyzed rice subgroups, especially between XI and GJ (Fig. 1b).
The GPC varied considerably between XI and GJ, with obvious regional differences. There is a high demand for rice varieties with grains that are rich in energy and nutrients among consumers in developing countries (e.g., in South and Southeast Asia), where XI varieties are commonly cultivated. In contrast, improving the rice grain quality is increasingly becoming a high priority among consumers in developed countries (e.g., in East Asia), where GJ varieties are predominant. In the present study, we revealed that the GNC varied more in the XI subgroup than in the GJ subgroup (Fig. 1b and Additional file 1: Table S1), which is consistent with the results of earlier studies on GPC (Chen et al. 2018; Zhou et al. 2009). However, the mean GNC of XI accessions was significantly lower than that of GJ accessions (Fig. 1b), which contradicts the findings of the reported studies on GPC (Chen et al. 2018; Zhou et al. 2009). This discrepancy may be due to the differences in the analyzed sample populations among the studies. Interestingly, the GNC was lower in MV than in LAN in the GJ subgroup, but not in the XI subgroup, and the GNC in MV was similar between the XI and GJ subgroups (Fig. 1c). These observations are suggestive of a stronger directional selection for the GNC in GJ than in XI. In other words, the GNC (or GPC) in modern GJ varieties was decreased during breeding to improve eating and cooking qualities.
Rice starch quality, which varies considerably between GJ and XI accessions, greatly influences rice cooking and processing methods for food and industrial applications (Umemoto et al. 1999). The substantial difference in the resistance to starch disintegration between GJ and XI is attributed to the diversity in the fine structures of the amylopectin in starch granules (Nakamura et al. 2002). Starch comprises 90% of the total dry weight of milled rice, and the amylose content is considered to be the most important factor affecting eating quality (Pang et al. 2016). In the present study, the GCC was significantly higher in MV than in LAN in the XI subgroup, whereas a similar significant difference in the GCC was not detected in the GJ subgroup (Fig. 1c). These results imply that increasing grain yield was a greater priority for XI varieties than for GJ varieties among breeding programs.
Comparisons with the Previously Reported Genes Related to GPC or GSC
The GPC- and GSC-related known genes near the QTLs identified in this study provide valuable information for thoroughly elucidating the putative genetic mechanisms underlying the GNC and GCC in rice. Although a number of QTLs for the GPC have been identified in rice germplasm (Liu et al. 2010; Ye et al. 2010; Zheng et al. 2011, 2012), relatively few have been cloned. A previous study proved that several mutations in a few genes have minor effects on the GPC and amino acid composition (Kawakatsu et al. 2010a). In the current study, some of the QTLs were only identified in XI or GJ. For example, a single-locus GWAS revealed four (qCN-1.2, qCN-7.1, qCN-9.1, and qCN-11.1) and three (qCN-3.1, qCN-4.1, and qCN-5.2) QTLs for the C/N ratio that were exclusive to the GJ and XI subgroups, respectively (Table 1). Moreover, OsAPP6 expression is reportedly associated with GPC variations only in XI rice (Peng et al. 2014). A recent study indicated that OsGluA2LET and OsGluA2HET, which are two OsGluA2 haplotypes, are present mainly in GJ and XI varieties, respectively (Yang et al. 2019). Thus, our results provide further evidence that the differences in the GPC between XI and GJ varieties depend on the diversity in the genetic architecture (Shi et al. 1999).
We searched the Oryzabase online resource (https://shigen.nig.ac.jp/rice/oryzabase/) for known rice genes that co-localized with the 16 QTLs identified in our single-locus GWAS. None of the identified genes are directly related to the GPC or GSC. When we extended the search to regions adjacent to these QTLs (within 100 kb), three known genes (CCRT, OsBZR1, and OsPPDKB) related to the GPC or GSC were detected. Regarding the GCC, qCC-5.2, with the most significant associated SNP (rs5_29562689, P = 7.43 × 10− 8) in the whole population panel, was detected close (approximately 76 kb downstream) to the starch synthesis-related gene CCRT (Table 1). A previous study proved that CCRT, which positively regulates starch synthesis in rice vegetative organs, is responsive to the photosynthate content and co-regulates the expression of rice genes related to starch synthesis (Morita et al. 2015). The C/N ratio is significantly and positively correlated with rice grain yield (Ye et al. 2014). In the current study, qCN-7.1, with a suggestive association (rs7_23515974, P = 2.99 × 10− 6) in the GJ panel, was detected approximately 30 kb from OsBZR1, which encodes a BR-signaling factor. The overexpression of OsBZR1 can enhance sugar accumulation and increase the grain yield. Knocking down this gene decreases the rice grain weight and starch accumulation. During the pollen and grain development in rice, OsBZR1 can directly promote CSA expression, which directly leads to the expression of genes related to sugar distribution and metabolism (Zhu et al. 2015). Additionally, OsPPDKB, which encodes a regulator of carbon metabolism (Kang et al. 2005), is located about 56 kb downstream from qCC-5.1 and qCN-5.1 (19,481,277–19,681,277 bp) on chromosome 5. Moreover, OsPPDKB regulates the carbon flow associated with starch and fat biosynthesis during the grain-filling period. Compared with the wild-type control, the floury endosperm-4 mutant generated via the insertion of a T-DNA into OsPPDKB has a significantly higher fat content, a slightly higher GPC, and a similar GCC (Kang et al. 2005).
Another three known genes related to the GPC and GCC were detected near the significantly associated SNPs in a multi-locus GWAS. Specifically, GIF1, which encodes a cell wall invertase required for carbon partitioning during the early grain-filling period (Wang et al. 2008), is located near the GCC-associated SNP rs4_20223533 in the GJ panel. Additionally, PFPβ, which regulates carbon metabolism during the rice grain-filling period (Duan et al. 2016), and OsAlaAT1, which is essential for the regulation of starch storage in rice endosperm (Yang et al. 2015), were respectively detected near the significant SNPs rs6_7739418 and rs10_13054571 associated with the C/N ratio in the whole panel. These results provide insights into the genetic basis of the variations in the GNC and GCC involving multiple QTLs/genes.
Utility of the Favorable Haplotypes of Candidate Genes
An apparent strength of the GWAS is that it is convenient for identifying favorable alleles/haplotypes at associated loci in a large set of natural populations and for screening for appropriate germplasm carrying the target alleles/haplotypes for the subsequent breeding of new varieties. According to our results, a method combining a single-locus and a multi-locus GWAS is more powerful than classical bi-parental linkage mapping methods for identifying QTLs for complex traits controlled by multiple genes. The QTLs and the representative haplotypes of the candidate genes described herein (Additional file 6: Table S6) may be useful for future gene cloning and molecular breeding aimed at rapidly improving the GNC and GCC in rice.
The synthesis of starch from sugar (photosynthates/carbohydrates) consumes less energy than the synthesis of other rice grain components, and is conducive to dry-matter accumulation and high yield. Additionally, specific rice-based products, including rice flour (for noodles), rice syrup, and feed rice, require grains that differ in terms of the GSC and GPC. Rice is an important source of nutrition for people and animals (livestock and poultry) in developing countries. Thus, one strategy for breeding high-yielding varieties with a high GPC involves applying marker-assisted selection to pyramid the representative high-GCC, high-GPC, and low-C/N alleles/haplotypes for LOC_Os01g06240 (Hap6 and Hap5) at qNC-1.1, LOC_Os05g33300 (Hap2 and Hap3) at qCC-5.1, LOC_Os01g04360 (Hap2) at qCN-1.1, and LOC_Os05g43880 (Hap4) at qCN-5.2 (Figs. 3, 4, 5 and 6 and Additional file 7: Table S7). However, there is also a demand in developed countries for rice varieties with grains that have a relatively low GPC, which enhances the taste. To satisfy this demand, rice breeders should focus on applying representative high-GPC and high-C/N alleles/haplotypes for LOC_Os01g06240 (Hap4) at qNC-1.1, LOC_Os01g04360 (Hap5 and Hap1) at qCN-1.1, and LOC_Os05g43880 (Hap6 and Hap3) at qCN-5.2 (Figs. 4, 5 and 6 and Additional file 7: Table S7). Furthermore, the effects of stacking favorable alleles/haplotypes at these loci will need to be investigated.
Conclusions
The QTLs for the GNC, GCC, and C/N ratio identified in this study may be useful for clarifying the molecular mechanism underlying the GNC and GCC. Our findings may also be relevant for enhancing the application of the favorable haplotypes of candidate genes during the molecular breeding of new rice varieties that satisfy the diverse demands for the GNC and GCC.
Methods
Rice Germplasm and Evaluation of the GNC and GCC in Milled Rice
The GNC and GCC of the milled rice grains of 751 accessions with appropriate and similar heading dates from the 3K RGP (3K RGP 2014) (Additional file 1: Table S1) were evaluated. On the basis of the known population structure and division of subpopulations (Wang et al. 2018), the 751 accessions comprised 475 XI accessions (73 XI-1A, 29 XI-1B, 74 XI-2, 113 XI-3, and 186 XI-adm), 231 GJ accessions (136 GJ-tmp, 35 GJ-sbtrp, 35 GJ-trp, and 25 GJ-adm), 27 Aus accessions, 6 Bas accessions, and 12 admixture (adm) accessions. Accessions were planted in Sanya, China in 2018 and 28-day-old seedlings were transplanted to field plots with three rows of eight plants (20 × 17 cm spacing) for each accession. Two replicates were prepared for each accession. The management of the field plots followed normal local agricultural practices.
At maturity, five plants in the middle of the second row were harvested and bulked for each replicate of every accession. The grains of each accession were threshed and air-dried in a greenhouse. When the moisture content of the grains reached 13%, the samples were prepared for the subsequent analyses as follows. After milling and crushing, the grain samples were passed through a 100-mesh sieve, after which 80 mg rice flour was placed in a tin paper cylinder, wrapped, and pressed to form a medicinal tablet shape. The GNC and GCC of the milled rice samples were analyzed with the vario MACRO cube (Elementar Co., Hanau, Germany), which is based on the Dumas combustion method. During the measurements, the working temperatures of the combustion tube and reduction tube were set at 1150 °C and 850 °C in the CNS mode, respectively. The helium intake pressure was set at 1200–1250 mbar and the flow rate was approximately 600 ml/min. The mean trait values for the two replicates were used for the GWAS.
Statistical Analyses of Phenotypic Data
Differences in the mean GNC, GCC, and C/N ratio among the rice subgroups were evaluated by a one-way ANOVA and Duncan’s multiple mean comparison test (5% significance level), which were completed with the agricolae package in R. Correlation analyses of the three traits were conducted with the corrplot package in R.
Single-Locus GWAS
The 3K RGP 4.8mio SNP dataset was downloaded from the Rice SNP-Seek Database (http://snp-seek.irri.org/) (Alexandrov et al. 2015). The 2,994,907, 2,118,326, and 1,318,493 SNPs with minor allele frequencies > 5% and a missing data rate < 0.1 filtered by PLINK (Purcell et al. 2007) for the whole population, XI, and GJ panels, respectively, were used for the subsequent association analyses (Additional file 3: Table S3). The single-locus GWAS was completed with EMMAX (Kang et al. 2010) to determine the associations between each SNP and the GNC, GCC, and C/N ratio of milled rice. A Balding–Nichols matrix based on the pruned subset of genome-wide SNP data (with the ‘indep-pairwise 50 10 0.1’ parameter in PLINK) was used to create the kinship matrix. We calculated the eigenvectors of the kinship matrix with GCTA (Yang et al. 2011) and then used the first three principal components as covariates to capture the variance due to the population structure. The effective number of independent markers (N) was calculated with the GEC software (Li et al. 2012) and suggestive P-value thresholds of association (1/N) were calculated (Additional file 3: Table S3). We identified the genes harboring or flanking the suggestively associated SNPs and functionally annotated them based on the Nipponbare reference genome IRGSP 1.0 (Kawahara et al. 2013). The Manhattan and quantile-quantile plots for the GWAS results were created with the R package qqman (Turner 2014). To detect independently associated regions, multiple suggestively associated SNPs located in one estimated LD block were clustered as one QTL region, and the SNP with the minimum P value in a cluster was considered as the lead SNP. Each LD block containing the detected SNPs was estimated with the ‘--blocks’ command in PLINK according to the block definition suggested by Gabriel et al. (2002).
Multi-Locus GWAS
The multi-locus GWAS was completed with the same genotypes and phenotypes used for the single-locus GWAS and the multi-locus random-SNP-effect mixed linear model (mrMLM) (Wang et al. 2016) of the mrMLM package (https://cran.r-project.org/web/packages/mrMLM/index.html) in R. A critical LOD score of 3.0 was used for identifying significantly associated SNPs.
Haplotype Analysis of Candidate Genes
The haplotypes of all 239 genes annotated based on the Nipponbare reference genome IRGSP 1.0 (Kawahara et al. 2013) and located within the 16 detected QTLs in the single-locus GWAS were classified according to all SNPs within the coding sequence region of one gene in the 751 rice accessions. The KEGG pathways associated with these genes were determined with EXPath 2.0 (Chien et al. 2015). Haplotypes in at least 10 rice accessions were used for a phenotypic comparative analysis. A one-way ANOVA followed by Duncan’s test were completed with the agricolae package in R to screen for candidate genes. Four representative candidate genes were selected for a comprehensive analysis based on the intensity of the association signals in the single-locus GWAS, the significance of the haplotype analyses (ANOVA), the biochemically related functions, and the expression profiles. Two-sided Fisher’s exact tests in R were used to compare haplotype frequencies between the rice XI and GJ subgroups. Nucleotide diversity (π) and Tajima’s D value for each 10-kb window across the genome, with an overlapping 5-kb step size, were calculated for the 600-kb region flanking the candidate genes with the Variscan program (version 2.0.3) (Vilella et al. 2005). Gene expression profiles were downloaded from a rice expression profile database [RiceXPro (version 3.0)] (Sato et al. 2013).
Supplementary information
Additional file 1 : Table S1. Summary of 751 rice accessions and the GNC, GCC, and C/N ratio of the milled rice
Additional file 2 : Table S2. Correlations among traits observed in different rice subgroups
Additional file 3 : Table S3. Filtered and effective number of single nucleotide polymorphisms in each GWAS panel and adjusted significant P-value thresholds based on a Bonferroni correction
Additional file 4 : Table S4. List of 55 significant SNPs associated with the GNC, GCC, and C/N ratio of milled rice detected in a single-locus GWAS
Additional file 5 : Table S5. Association signals detected in a multi-locus GWAS for the GNC, GCC, and C/N ratio of milled rice
Additional file 6 : Table S6. Comparison of the GNC, GCC, and C/N ratio among the haplotypes of 239 annotated genes within 16 QTLs detected in a single-locus GWAS
Additional file 7 : Table S7. Haplotype analysis of four representative candidate genes at four QTLs associated with the GNC, GCC, and C/N ratio of milled rice
Acknowledgments
We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Abbreviations
- 3K RGP
3000 Rice Genomes Project
- adm
Admixture
- Bas
Basmati
- BE
Starch branching enzyme
- C
Carbon
- GJ
Geng/Japonica
- GNC
Grain N content
- GPC
Grain protein content
- GSC
Grain starch content
- GWAS
Genome-wide association study
- LAN
Landraces
- LD
Linkage disequilibrium
- MV
Modern varieties
- N
Nitrogen
- PVE
Phenotypic variation explained
- QTL
Quantitative trait locus
- SNP
Single nucleotide polymorphism
- SS
Soluble starch synthase
- SSP
Seed storage protein
- XI
Xian/Indica
- π
Nucleotide diversity
Authors’ Contributions
Conceived and designed the experiments: LT and FZ; Performed the experiments: AL, JS, SM, XW, ZL, WL, QL, and SC; Analyzed the data: FZ; Wrote the paper: LT and FZ. All authors read and approved the final manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2016YFD0300504, 2017YFD0100501), LiaoNing Revitalization Talents Program (XLYC1807233), Program for Liaoning Excellent Talents in University (LR2017007).
Availability of Data and Materials
All data supporting the conclusions of this article are provided within the article (and in the Additional files).
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liang Tang, Email: tangliang@syau.edu.cn.
Fan Zhang, Email: zhangfan03@caas.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12284-019-0362-2.
References
- 3K RGP The 3,000 rice genomes project. Gigascience. 2014;3:7. doi: 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z, Mauleon R, Hamilton RS, McNally KL. SNP-seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beljkaš B, Matić J, Milovanović I, Jovanov P, Mišan A, Šarić L. Rapid method for determination of protein content in cereals and oilseeds: validation measurement uncertainty and comparison with the Kjeldahl method. Accred Qual Assur. 2010;15:555–561. doi: 10.1007/s00769-010-0677-6. [DOI] [Google Scholar]
- Chen P, Shen Z, Ming L, Li Y, Dan W, Lou G, Peng B, Wu B, Li Y, Zhao D, Gao G, Zhang Q, Xiao J, Li X, Wang G, He Y. Genetic basis of bariation in rice seed storage protein (albumin globulin prolamin and glutelin) content revealed by genome-wide association analysis. Front Plant Sci. 2018;9:612. doi: 10.3389/fpls.2018.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Xu Q, Zheng T, Ye G, Luo C, Xu J, Li Z. Identification of stably expressed quantitative trait loci for grain yield and protein content using recombinant inbred line and reciprocal introgression line populations in rice. Crop Sci. 2013;53:1437–1446. doi: 10.2135/cropsci2013.02.0075. [DOI] [Google Scholar]
- Chien C, Chow C, Wu N, Chiang-Hsieh Y, Hou P, Chang W. EXPath: a database of comparative expression analysis inferring metabolic pathways for plants. BMC Genomics. 2015;16(Suppl 2):S6. doi: 10.1186/1471-2164-16-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F, Wang L, Mei XF, Li SX, Pu SL, Li QP, Ren WJ. Polyaspartic acid (PASP)-urea and optimised nitrogen management increase the grain nitrogen concentration of rice. Sci Rep. 2019;9:313. doi: 10.1038/s41598-018-36371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan E, Wang Y, Liu L, Zhu J, Zhong M, Zhang H, Li S, Ding B, Zhang X, Guo X, Jiang L, Wan J. Pyrophosphate: fructose-6-phosphate 1-phosphotransferase (PFP) regulates carbon metabolism during grain filling in rice. Plant Cell Rep. 2016;35:1321–1331. doi: 10.1007/s00299-016-1964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Isabella B, Ines H, Michael R. Determination of total organic carbon – an overview of current methods. Trends Anal Chem. 2004;23:716–726. doi: 10.1016/j.trac.2004.09.003. [DOI] [Google Scholar]
- Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y. Starch biosynthesis in cereal endosperm. Plant Physiol Biochem. 2010;48:383–392. doi: 10.1016/j.plaphy.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Kang HG, Park S, Matsuoka M, An G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB) Plant J. 2005;42:901–911. doi: 10.1111/j.1365-313X.2005.02423.x. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Hirose S, Yasuda H, Takaiwa F. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 2010;154:1842–1854. doi: 10.1104/pp.110.164343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Takaiwa F. Cereal seed storage protein synthesis: fundamental processes for recombinant protein production in cereal grains. Plant Biotechnol J. 2010;8:939–953. doi: 10.1111/j.1467-7652.2010.00559.x. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Hirose S, Yano M, Takaiwa F. Characterization of a new rice glutelin gene glud-1 expressed in the starchy endosperm. J Exp Bot. 2008;59:4233–4245. doi: 10.1093/jxb/ern265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Touno SM, Yasuda H, Takaiwa F. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 2010;59:908–920. doi: 10.1111/j.1365-313X.2009.03925.x. [DOI] [PubMed] [Google Scholar]
- Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wan X, Ma X, Wan J. Dissecting the genetic basis for the effect of rice chalkiness amylose content protein content and rapid viscosity analyzer profile characteristics on the eating quality of cooked rice using the chromosome segment substitution line population across eight environments. Genome. 2010;54:64–80. doi: 10.1139/G10-070. [DOI] [PubMed] [Google Scholar]
- Long X, Liu Q, Chan M, Wang Q, Sun SS. Metabolic engineering and profiling of rice with increased lysine. Plant Biotechnol J. 2013;11:490–501. doi: 10.1111/pbi.12037. [DOI] [PubMed] [Google Scholar]
- Lu FH, Park YJ. An SNP downstream of the OsBEIIb gene is significantly associated with amylose content and viscosity properties in rice (Oryza sativa L.) J Cereal Sci. 2012;56:706–712. doi: 10.1016/j.jcs.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Makoto K, Kenzo M, Shuichi I, Hiroyuki F, Toshiya T, Hidenori S, Minoru N, Takeshi N. Low glutelin content1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell. 2003;15:1455–1467. doi: 10.1105/tpc.011452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Fitzgerald MA. Proteins in rice grains influence cooking properties! J Cereal Sci. 2002;36:285–294. doi: 10.1006/jcrs.2001.0465. [DOI] [Google Scholar]
- Morita R, Sugino M, Hatanaka T, Misoo S, Fukayama H. CO2-responsive CONSTANS CONSTANS-like and time of chlorophyll a/b binding protein Expression1 protein is a positive regulator of starch synthesis in vegetative organs of rice. Plant Physiol. 2015;167:1321–1331. doi: 10.1104/pp.15.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Sakurai A, Inaba Y, Kimura K, Iwasawa N, Nagamine T. The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. Starch - Stärke. 2002;54:117–131. doi: 10.1002/1521-379X(200204)54:3/4<117::AID-STAR117>3.0.CO;2-2. [DOI] [Google Scholar]
- Ning H, Qiao J, Liu Z, Lin Z, Li G, Wang Q, Wang S, Ding Y. Distribution of proteins and amino acids in milled and brown rice as affected by nitrogen fertilization and genotype. J Cereal Sci. 2010;52:90–95. doi: 10.1016/j.jcs.2010.03.009. [DOI] [Google Scholar]
- Pang Y, Ali J, Wang X, Franje NJ, Revilleza JE, Xu J, Li Z. Relationship of rice grain amylose gelatinization temperature and pasting properties for breeding better eating and cooking quality of rice varieties. PLoS One. 2016;11:e0168483. doi: 10.1371/journal.pone.0168483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Kong H, Li Y, Wang L, Zhong M, Sun L, Gao G, Zhang Q, Luo L, Wang G, Xie W, Chen J, Yao W, Peng Y, Lei L, Lian X, Xiao J, Xu C, Li X, He Y. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat Commun. 2014;5:4847. doi: 10.1038/ncomms5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Wang Y, Liu F, Zhou K, Ding Y, Feng Z, Wang Y, Liu K, Gan L, Ma W. GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell. 2014;26:410–425. doi: 10.1105/tpc.113.121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Shigemitsu T, Yamasaki R, Sasou A, Goto F, Kishida K, Kuroda M, Tanaka K, Morita S, Satoh S. Formation mechanism of the internal structure of type I protein bodies in rice endosperm: relationship between the localization of prolamin species and the expression of individual genes. Plant J. 2012;70:1043–1055. doi: 10.1111/j.1365-313X.2012.04947.x. [DOI] [PubMed] [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio B, Nagamura Y. RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013;41:D1206–D1213. doi: 10.1093/nar/gks1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She K, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell. 2010;22:3280–3294. doi: 10.1105/tpc.109.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Zhu J, Yang X, Yu Y, Wu J. Genetic analysis for protein content in indica rice. Euphytica. 1999;107:135–140. doi: 10.1023/A:1026435323139. [DOI] [Google Scholar]
- Shin JH, Blay S, McNeney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw. 2006;16:9. doi: 10.18637/jss.v016.c03. [DOI] [Google Scholar]
- Tang L, Xu ZJ, Chen WF. Advances and prospects of super rice breeding in China. J Integr Agric. 2017;16:984–991. doi: 10.1016/S2095-3119(16)61604-0. [DOI] [Google Scholar]
- Tang XJ, Peng C, Zhang J, Cai Y, You XM, Kong F, Yan HG, Wang GX, Wang L, Jin J, Chen WW, Chen XG, Ma J, Wang P, Jiang L, Zhang WW, Wan JM. ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Sci. 2016;249:70–83. doi: 10.1016/j.plantsci.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Terao T, Hirose T. Control of grain protein contents through SEMIDWARF1 mutant alleles: sd1 increases the grain protein content in Dee-geo-woo-gen but not in Reimei. Mol Gen Genomics. 2015;290:939–954. doi: 10.1007/s00438-014-0965-7. [DOI] [PubMed] [Google Scholar]
- Tian L, Ling LD, Zhi JY, Fukuda M, Kumamaru T, Xiang BD, Xiu PX, Le QQ. Small GTPase Sar1 is crucial for proglutelin and α-globulin export from the endoplasmic reticulum in rice endosperm. J Exp Bot. 2013;64:2831–2845. doi: 10.1093/jxb/ert128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Qian Q, Liu Q, Yan M, Liu X, Yan C, Liu G, Gao Z, Tang S, Zeng D. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci U S A. 2009;106:21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD (2014) Qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. bioRxiv. 10.1101/005165
- Umemoto T, Terashima K, Nakamura Y, Satoh H. Differences in amylopectin structure between two rice varieties in relation to the effects of temperature during grain-filling. Starch - Stärke. 1999;51:58–62. doi: 10.1002/(SICI)1521-379X(199903)51:2<58::AID-STAR58>3.0.CO;2-J. [DOI] [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, Ma H, Zhang G, He Z. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- Wang G, Sun X, Wang G, Wang F, Gao Q, Sun X, Tang Y, Chang C, Lai J, Zhu L. Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics. 2011;189:1281–1295. doi: 10.1534/genetics.111.133967. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang SB, Feng JY, Ren WL, Huang B, Zhou L, Wen YJ, Zhang J, Dunwell JM, Xu S, Zhang YM. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci Rep. 2016;6:19444. doi: 10.1038/srep19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis KC, Xu J, Sun C, Fu B, Zhang H, Gao Y, Zhao X, Shen F, Cui X, Yu H, Li Z, Chen M, Detras J, Zhou Y, Zhang X, Zhao Y, Kudrna D, Wang C, Li R, Jia B, Lu J, He X, Dong Z, Xu J, Li Y, Wang M, Shi J, Li J, Zhang D, Lee S, Hu W, Poliakov A, Dubchak I, Ulat VJ, Borja FN, Mendoza JR, Ali J, Li J, Gao Q, Niu Y, Yue Z, Naredo MEB, Talag J, Wang X, Li J, Fang X, Yin Y, Glaszmann JC, Zhang J, Li J, Hamilton RS, Wing RA, Ruan J, Zhang G, Wei C, Alexandrov N, McNally KL, Li Z, Leung H. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pang Y, Zhang J, Wu Z, Chen K, Ali J, Ye G, Xu J, Li Z. Genome-wide and gene-based association mapping for rice eating and cooking characteristics and protein content. Sci Rep. 2017;7:17203. doi: 10.1038/s41598-017-17347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Liu X, Jiang L, Chen L, Han X, Jin M, Liu S, Liu F, Lv J. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J Cell Mol Biol. 2010;64:812–824. doi: 10.1111/j.1365-313X.2010.04370.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu S, Liu S, Jiang L, Chen L, Ren Y, Han X, Liu F, Ji S, Liu X. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 2010;58:606–617. doi: 10.1111/j.1365-313X.2009.03801.x. [DOI] [PubMed] [Google Scholar]
- Wu CY, Washida H, Onodera Y, Harada K, Takaiwa F. Quantitative nature of the prolamin-box ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. Plant J Cell Mol Biol. 2010;23:415–421. doi: 10.1046/j.1365-313x.2000.00797.x. [DOI] [PubMed] [Google Scholar]
- Xu F, Bao J, He Q, Park YJ. Genome-wide association study of eating and cooking qualities in different subpopulations of rice (Oryza sativa L.) BMC Genomics. 2016;17:663. doi: 10.1186/s12864-016-3000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang G, Yang X, Li Z, Feng H, Xu B, Zhao X. Monitoring ratio of carbon to nitrogen (C/N) in wheat and barley leaves by using spectral slope features with branch-and-bound algorithm. Sci Rep. 2018;8:10034. doi: 10.1038/s41598-018-28351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kim SR, Lee SK, Choi H, Jeon JS, An G. Alanine aminotransferase 1 (OsAlaAT1) plays an essential role in the regulation of starch storage in rice endosperm. Plant Sci. 2015;240:79–89. doi: 10.1016/j.plantsci.2015.07.027. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Sun C, Bai J, Luo Z, Shi B, Zhang J, Yan W, Piao Z. A putative gene sbe3-rs for resistant starch mutated from SBE3 for starch branching enzyme in rice (Oryza sativa L.) PLoS One. 2012;7:e43026. doi: 10.1371/journal.pone.0043026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo M, Sun S, Zou Y, Yin S, Liu Y, Tang S, Gu M, Yang Z, Yan C. Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat Commun. 2019;10:1949. doi: 10.1038/s41467-019-09919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Okuno K, Satoh H, Omura T. Chromosomal location of genes conditioning low amylose content of endosperm starches in rice Oryza sativa L. Theor Appl Genet. 1988;76:183–189. doi: 10.1007/BF00257844. [DOI] [PubMed] [Google Scholar]
- Yao X, Wang J, Jin L, Jia Z, Ren C, Ma D, Hai X, Xu Z. Mapping quantitative trait loci associated with starch paste viscosity in rice ( Oryza sativa L.) under different environmental conditions. Plant Breed. 2017;136:591–602. doi: 10.1111/pbr.12505. [DOI] [Google Scholar]
- Ye G, Liang S, Wan J. QTL mapping of protein content in rice using single chromosome segment substitution lines. Theor Appl Genet. 2010;121:741–750. doi: 10.1007/s00122-010-1345-2. [DOI] [PubMed] [Google Scholar]
- Ye Y, Liang X, Chen Y, Li L, Ji Y, Zhu C. Carbon, nitrogen and phosphorus accumulation and partitioning, and C:N:P stoichiometry in late-season rice under different water and mitrogen managements. PLoS One. 2014;9:e101776. doi: 10.1371/journal.pone.0101776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Aijun S, Mei W, Zhen Z, Ouwerkerk PBF. Functions of the CCCH type zinc finger protein OsGZF1 in regulation of the seed storage protein GluB-1 from rice. Plant Mol Biol. 2014;84:621–634. doi: 10.1007/s11103-013-0158-5. [DOI] [PubMed] [Google Scholar]
- Zhan X, Shen Q, Wang X, Hong Y. The sulfoquinovosyltransferase-like enzyme SQD2.2 is involved in flavonoid glycosylation, regulating sugar metabolism and seed setting in rice. Sci Rep. 2017;7:4685. doi: 10.1038/s41598-017-04002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QF, Zhang YD, Zhu Z, Zhao L, Zhao QY, Ling XU, Wang CL. Inheritance analysis and QTL mapping of rice starch viscosity (rapid visco analyzer profile) characteristics. Rice Sci. 2008;15:186–194. doi: 10.1016/S1672-6308(08)60041-4. [DOI] [Google Scholar]
- Zheng L, Zhang W, Chen X, Jing M, Chen W, Zhao Z, Zhai H, Wan J. Dynamic QTL analysis of rice protein content and protein index using recombinant inbred lines. J Plant Biol. 2011;54:321–328. doi: 10.1007/s12374-011-9170-y. [DOI] [Google Scholar]
- Zheng L, Zhang W, Liu S, Chen L, Liu X, Chen X, Ma J, Chen W, Zhao Z, Jiang L, Wan J. Genetic relationship between grain chalkiness protein content and paste viscosity properties in a backcross inbred population of rice. J Cereal Sci. 2012;56:153–160. doi: 10.1016/j.jcs.2012.05.003. [DOI] [Google Scholar]
- Zhou H, Wang L, Liu G, Meng X, Jing Y, Shu X, Kong X, Sun J, Yu H, Smith SM. Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase waxy in synthesizing resistant starch in rice. Proc Natl Acad Sci U S A. 2016;113:12844–12849. doi: 10.1073/pnas.1615104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LH, Liu QQ, Changquan Z, Xu Y, Tang SZ, Gu MH. Variation and distribution of seed storage protein content and composition among different rice varieties. Acta Agron Sin. 2009;35:884–891. doi: 10.3724/SP.J.1006.2009.00884. [DOI] [Google Scholar]
- Zhu X, Liang W, Cui X, Chen M, Yin C, Luo Z, Zhu J, Lucas WJ, Wang Z, Zhang D. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of carbon starved anther a MYB domain protein. Plant J. 2015;82:570–581. doi: 10.1111/tpj.12820. [DOI] [PubMed] [Google Scholar]
- Vilella A. J., Blanco-Garcia A., Hutter S., Rozas J. VariScan: Analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21(11):2791–2793. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 : Table S1. Summary of 751 rice accessions and the GNC, GCC, and C/N ratio of the milled rice
Additional file 2 : Table S2. Correlations among traits observed in different rice subgroups
Additional file 3 : Table S3. Filtered and effective number of single nucleotide polymorphisms in each GWAS panel and adjusted significant P-value thresholds based on a Bonferroni correction
Additional file 4 : Table S4. List of 55 significant SNPs associated with the GNC, GCC, and C/N ratio of milled rice detected in a single-locus GWAS
Additional file 5 : Table S5. Association signals detected in a multi-locus GWAS for the GNC, GCC, and C/N ratio of milled rice
Additional file 6 : Table S6. Comparison of the GNC, GCC, and C/N ratio among the haplotypes of 239 annotated genes within 16 QTLs detected in a single-locus GWAS
Additional file 7 : Table S7. Haplotype analysis of four representative candidate genes at four QTLs associated with the GNC, GCC, and C/N ratio of milled rice
Data Availability Statement
All data supporting the conclusions of this article are provided within the article (and in the Additional files).