Figure 2.
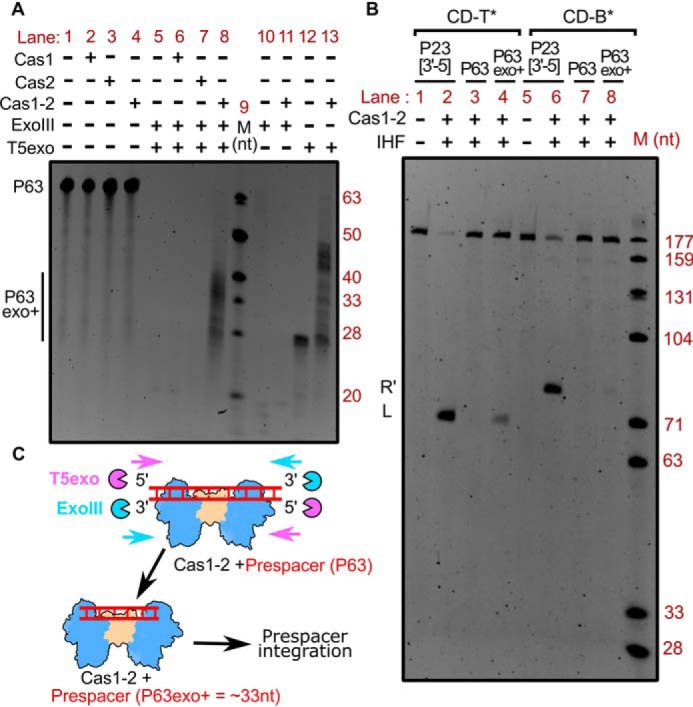
Tailoring of Cas1-2 bound large DNA fragments by exonucleases generates integration-competent prespacers. A, denaturing PAGE depicting the nuclease treatment of Cas1-2 bound P63 DNA fragments. The presence (+) or absence (−) of each reaction component is indicated at the top of each lane. Positions corresponding to the substrate (P63) and T5 exo/ExoIII-digested DNA fragments (P63exo+) are indicated on the left, whereas oligonucleotide marker (M) positions are shown on the right. B, denaturing PAGE displaying the integration reactions employing various prespacers (P23[3′-5] (lanes 1 and 2 and lanes 5 and 6), P63 (lanes 3 and 7), and Cas1-2 protected DNA fragments (P63exo+) (lanes 4 and 8)) and CRISPR DNA substrates (CD-T* (lanes 1–4) and CD-B* (lanes 5–8)) is shown. The presence (+) or absence (−) of each reaction component is indicated at the top of each lane. Positions corresponding to labeled DNA products that result from prespacer nucleophilic attack (L and R′) are displayed on the left. The DNA molecular weight marker (M) positions are shown on the right. C, schema illustrating the mechanism of Cas1-2–mediated protection of prespacer boundaries. Cas1-2 (in blue and brown), T5 exonuclease (magenta pie), exonuclease III (cyan pie), and prespacer P63 (red ladder) are portrayed.
