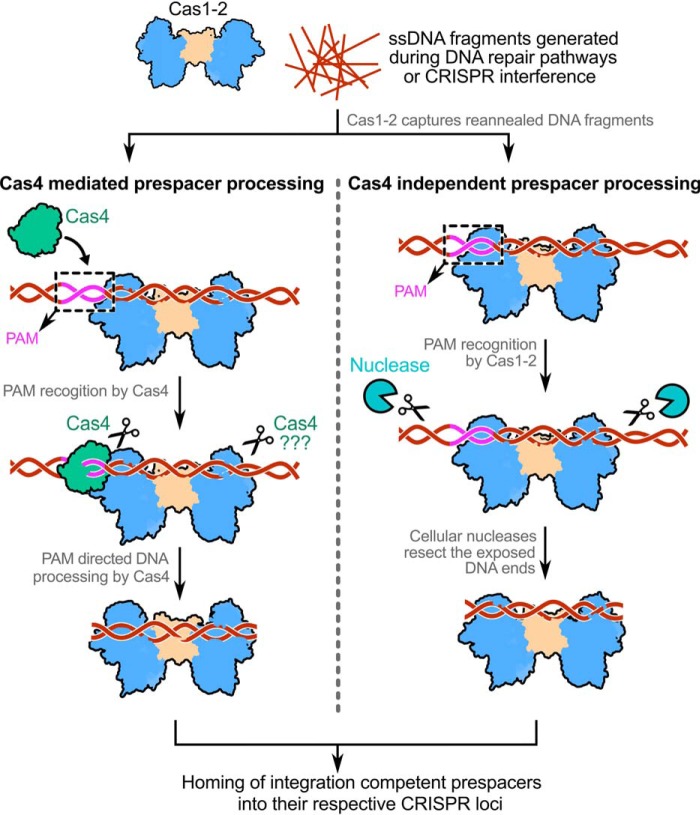
Figure 7.
Model depicting the mechanism of Cas4-dependent and -independent prespacer processing in type I CRISPR-Cas systems. The prespacer production in CRISPR systems encompasses a subset of two events: (i) PAM directed prespacer capture and (ii) processing of selected prespacer to a defined length for integration at the CRISPR locus. Generally, Cas1-2 captures long dsDNA fragments that are produced by reannealing of ssDNA products (in brown) derived from DNA repair pathways (such as RecBCD in E. coli) or CRISPR interference. Although in type I-E it is clear that the DNA capture by Cas1-2 precedes the prespacer processing event, the order of DNA capture and processing is yet to be understood in other CRISPR-Cas subtypes. The Cas4 (in green) in CRISPR-Cas subtypes I-A, I-C, I-D, and I-G or the Cas4 domain of Cas4-Cas1 fusion in type I-U trims the DNA upon recognizing the PAM region (in magenta) (25, 45–51). A second copy of Cas4 in type I-G was shown to trim the non-PAM end upon recognizing a short motif (47), whereas in other subtypes, it is not clear whether Cas4 processes this end. Here, the dual role of PAM recognition and prespacer processing by Cas4 propels CRISPR adaptation. Unlike this, in the type I-E system, the intrinsic affinity of Cas1-2 integrase toward PAM itself is sufficient to define the potential prespacer regions. This aspect of Cas1-2/I-E precludes the involvement of any PAM-specific Cas nucleases (such as Cas4) in prespacer selection. As the Cas1-2/I-E protects the prespacer boundaries efficiently upon recognizing the PAM, any common cellular non-Cas nucleases (in cyan) could trim the exposed ends to generate aptly sized prespacers for integration into the CRISPR locus.