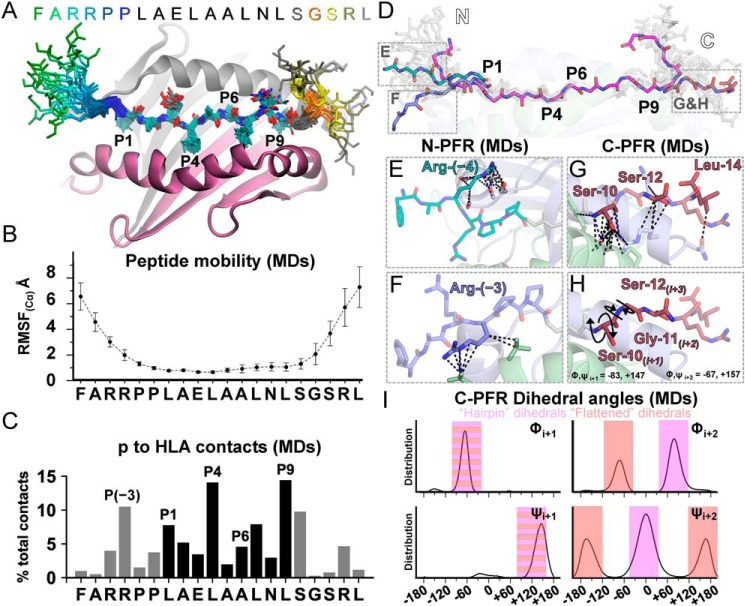
Figure 6.
Conformations explored by PFRs of 5T4111–130 in MD simulations. A, representative snapshots from the 20 most populated clusters of the MD trajectories illustrating peptide mobility. Backbone atoms of PFRs are shown as sticks and color-coded as indicated (inset). Core residues (all atoms) are blue sticks with atom coloring as previous. DRα (gray cartoon) and DR1β (purple cartoon) are represented from the static crystallographic structure. B, RMSF of peptide Cα atoms. Mean ± S.D. as extracted from 10 independent MD simulations is shown. C, percentage of the total number of peptide to HLA contacts (all-atoms including modeled hydrogens = <3.0 Å) calculated from the MDs. D, three highlighted alternative PFR conformations represented in MDs. Crystallographic peptide conformation is shown by pink sticks. Magnified visualization of boxed regions is shown in E-H with matched coloring accordingly. E, extension of contacts to Arg-(−4) through binding to -DRα residues observed in MD simulations. F, alternative conformation whereby Arg-(−3) interacted with -DRβ residues in MD simulations. G, representative extended conformation of Ser10–Leu14 extracted from MD simulations whereby a potential flattening of the C-PFR hairpin loop allows contact with HLA residues in MDs. H, this suggested flattened conformation was enabled by inversion of dihedral angles about Gly-11. I, distribution of backbone dihedral angles (ϕ, ψ) of residues Ser-10(i+1) and Gly-11(i+2) extracted from the MDs. Ideal angles describing the crystallographic hairpin loop and flattened conformation are boxed in pink and red, respectively (±40° deviation from ideals).