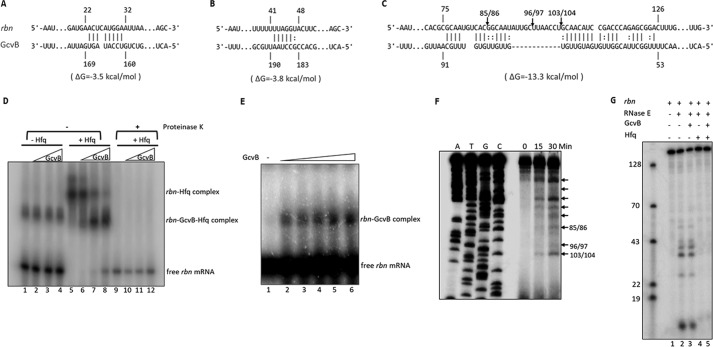
Figure 9.
In vitro interaction of GcvB with rbn mRNA and cleavage by purified RNase E. A–C, predicted base pairing between GcvB and three regions of the 5′ 165-nt fragment of rbn mRNA using the IntaRNA program. Numbering of nucleotides on rbn mRNA and GcvB is relative to the transcription start site of each RNA. Arrows indicate sites of cleavage by RNase E determined by the primer extension assay shown in E. D, interaction of GcvB with rbn mRNA. 5′-Labeled rbn fragment (0.1 pmol) and 0.4 pmol (lanes 2, 6, and 10), 0.8 pmol (lanes 3, 7, and 11), or 1.6 pmol (lanes 4, 8, and 12) of GcvB were incubated with or without purified Hfq in a 10-μl reaction. For reactions in lanes 9–12, 10 μg of proteinase K was added following complex formation to remove Hfq. Samples were analyzed on 5% native polyacrylamide gels. A representative experiment carried out twice with essentially identical results is shown. E, interaction of GcvB with rbn mRNA in the absence of Hfq. 5′-Labeled rbn fragment (1 pmol) was incubated with 1, 2, 4, 8, or 16 pmol of GcvB (lanes 2–6, respectively) in a 20-μl reaction. F, analysis of RNase E cleavage sites on rbn mRNA. The rbn fragment (1–165) was synthesized by in vitro transcription and digested with purified RNase E (N-terminal protein, 1–498). Digestion products were subjected to primer-extension analysis as described under “Experimental procedures.” Arrows indicate all time-dependent cleavage products. The positions of the three cleavage sites within the region of highest complementarity to GcvB are numbered based on the sequence ladders. G, effect of GcvB and Hfq on in vitro cleavage of rbn mRNA by RNase E. Reaction mixtures were set up as described under “Experimental procedures.” Reaction mixtures lacking RNase E were first incubated for 20 min at 37 °C, followed by addition of RNase E and additional incubation for 15 min. Cleavage products were analyzed by 8% urea-PAGE. A mixture of 5′-labeled DNA oligonucleotides was used to evaluate the size of cleavage products. D–G, representative experiments carried out twice with essentially identical results are shown.