Fig. 4.
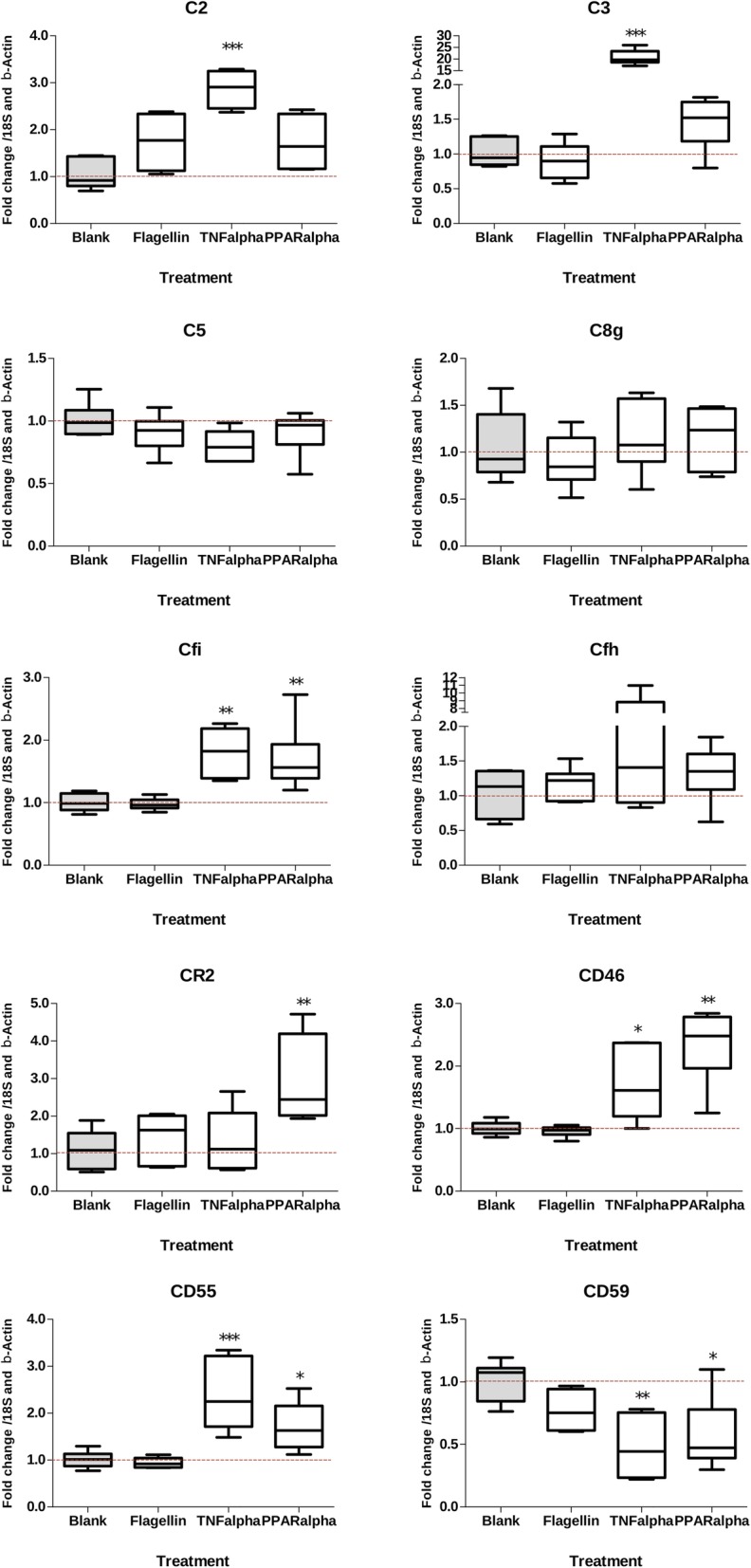
Expression of 10 chosen genes from the ‘Regulation of Complement Cascade’ pathways with significance calculated with ANOVA. Each graph contains information on different genes, the x-axis contains information on the treatment of the organoids and the y-axis has the fold change of the control genes. Data were analysed using Prism statistical software (v5.0, Graphpad, San Diego, US), measured for normality using the Kolmogorov-Smirnov test, and represented as Box and Whisker plots. A t-statistic test was performed on the RT-qPCR results of the 10 genes using the same methods as on the nodes in the gene set pathway analysis of CePa. All data were considered significantly different from the Blank (indicated in grey) when P < alpha (0.05) and indicated with * (P < 0.05 = *, P < 0.01 = **, P < 0.001 = ***)
