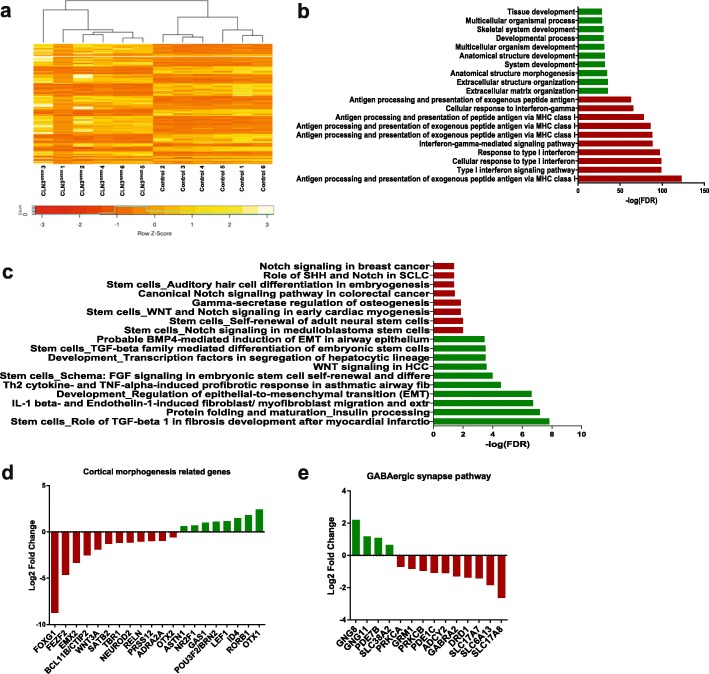
Fig. 5.
Whole-transcriptome analysis reveals impaired development in CLN3Q352X cerebral organoids. a Heatmap clustering the differentially expressed genes between Control (healthy) and CLN3Q352X (mutant) brain organoids. b Gene enrichment analysis of CLN3Q352X network. Genes that are upregulated in disease phenotype were significantly enriched in cellular processes highlighted in green, while downregulations are depicted in red. c Pathway enrichment analysis of the CLN3Q352X network. Pathway up-regulations are highlighted in green, while down-regulations are marked in red. d Log 2 Fold change expression values for genes related to brain development and cortical morphogenesis were generally decreased in CLN3Q352X. e Log 2 Fold change expression values for genes related to synapses, which were mostly downregulated in the CLN3Q352X organoids. Genes in D and E are specifically differentially in our dataset (but not necessarily present in the networks) and belong to the pathways extracted from Pathway unification database (PathCards)