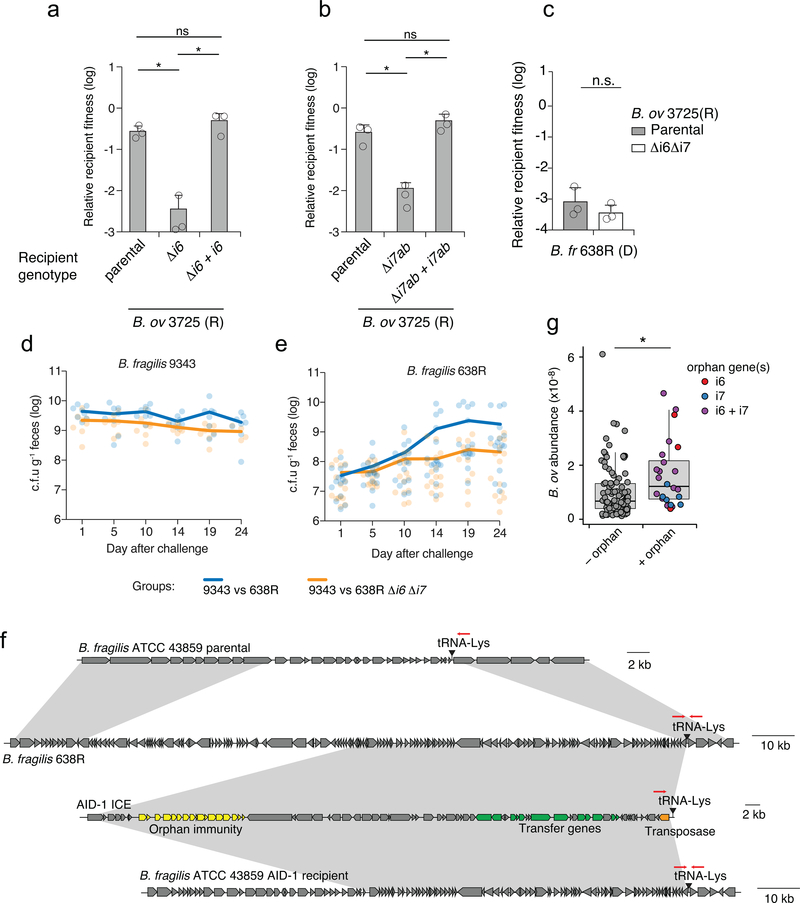
Extended Data Fig. 3 |. Orphan immunity genes specifically enhance the fitness of Bacteroides strains in vitro and in vivo.
a, b, T6SS-dependent competitiveness of parental strains of B. ovatus 3725 and the indicated mutant and complemented derivatives during in vitro growth competition experiments with B. fragilis 9343. Relative recipient fitness was determined by calculating the ratio of final to initial colony forming units and normalizing to the corresponding experiment with B. fragilis 9343 lacking tssC (T6S-inactive). Data represent mean ± s.d. of n=3 independent biological replicates, unpaired two-tailed t-test, *P < 0.01. c, T6SS-dependent competitiveness of a parental strain of B. ovatus 3725 or a strain bearing in-frame deletions of indicated orphan immunity genes, during in vitro growth competition experiments with an orthogonal effector-bearing B. fragilis 638R parental strain or a derivative strain lacking tssC (T6-inactive). Relative recipient fitness and statistics were calculated as in a, b, n=3 independent biological replicates. d, e, Recovery of B. fragilis 9343 d, or 638R and the indicated orphan immunity mutant derivative e, from pairwise competitions of the strains in germ-free mice. Lines indicate the mean at each time point, (n=8 mice per group for each of two independent experiments). Alternating time points of these data are included in ratio form in Fig. 3c. f, Schematic depicting genomic loci for the B. fragilis ATCC 43859 parental strain, the B. fragilis 638R AID-1 donor strain, the AID-1 system, and the ATCC 43859 AID-1 recipient. Grey shading indicates homology, red arrows indicate the position of PCR primers used to infer insertion of the AID-1 element at the tRNA-Lys insertion site. g, Abundance of B. ovatus in samples lacking detected orphan immunity genes (−) and samples in which the indicated orphan immunity genes were assigned to B. ovatus (+). Abundances are calculated as in Fig. 1a. (Wilcoxon rank sum test, *P<0.001, n=128 non-orphan samples and n=24 for samples harboring orphan immunity, box plots indicate interquartile range with line at the median and whiskers indicating 1.5 times the IQR).