Figure 1. Renewable barcoding system and lineage dynamics.
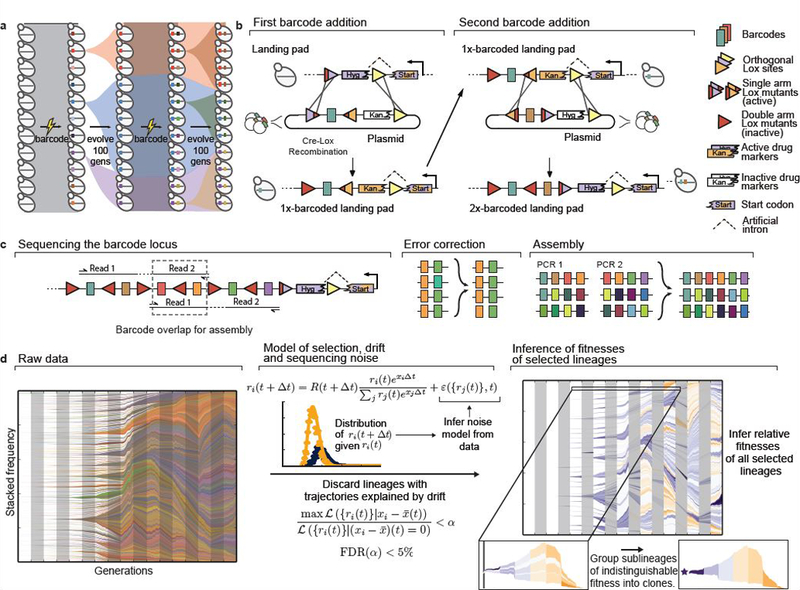
a, Experimental design. Diverse DNA barcodes are introduced into an initially clonal population; each barcode labels a small lineage. Every 100 generations, we introduce new diverse barcodes immediately adjacent to existing barcodes, subdividing each lineage into sublineages. b, Renewable barcoding system, using a novel Cre-Lox system consisting of three orthogonal Lox sites (colored triangles), each of which can be modified with two arm disruptions (red shading) that are individually tolerated but jointly inactivating (SI section 1). At each barcode addition, we combine arm disruptions to inactivate the old Lox site, while adding a new orthogonal active Lox site; alternating Lox orientations further limit undesired recombination. Drug markers contain an intron 3’ splice accepting site and must correctly integrate at the landing pad containing the 5’ splice donor to be functional. c, When the barcode locus exceeds the length of an Illumina read, we use custom priming sites to sequence overlapping sets of four consecutive barcodes. After exploiting barcode diversity to identify and correct sequencing errors, we use these overlaps to unambiguously reconstruct the full barcode locus (SI section 2). d, Inference pipeline. At left, raw barcode frequencies over time (left to right, colors chosen at random). For legibility, we only show lineages or sublineages whose frequency exceeds 0.1% in at least one timepoint; combined frequencies of lineages that do not individually reach 0.1% are shown as white space (or the color of the parent when that parent exceeds 0.1% frequency). Center panel summarizes model for identifying selected lineages. Briefly, we use the data to construct a parametric model for the strength of noise from genetic drift and sequencing and discard trajectories explained by noise alone. We then jointly infer fitnesses of all remaining lineages, and group lineages of indistinguishable fitness into clones.
