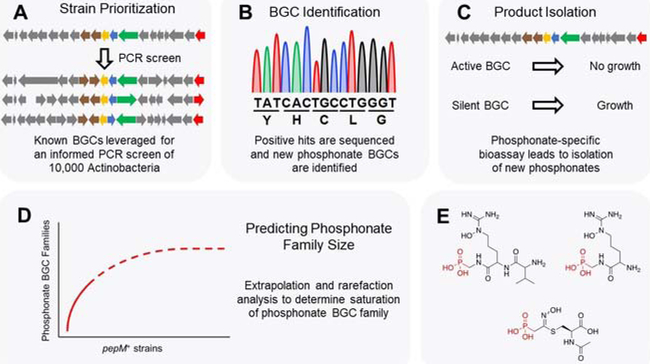
Figure 4: Genome mining of phosphonate NPs.
(A) A PCR screen targeting the pepM gene was used to identify phosphonate BGCs from 10,000 Actinobacteria. (B) Hit strains from the PCR screen were sequenced and phosphonate BGCs identified therein. (C) Extracts from strains containing phosphonate BGCs were assayed with an engineered E. coli strain hypersensitive to phosphonates. (D) Bioinformatics and statistics were used to map the diversity of phosphonate BGCs and to extrapolate how much phosphonate diversity remains in Actinobacteria. (E) Examples of novel phosphonate compounds isolated from genome mining are shown.