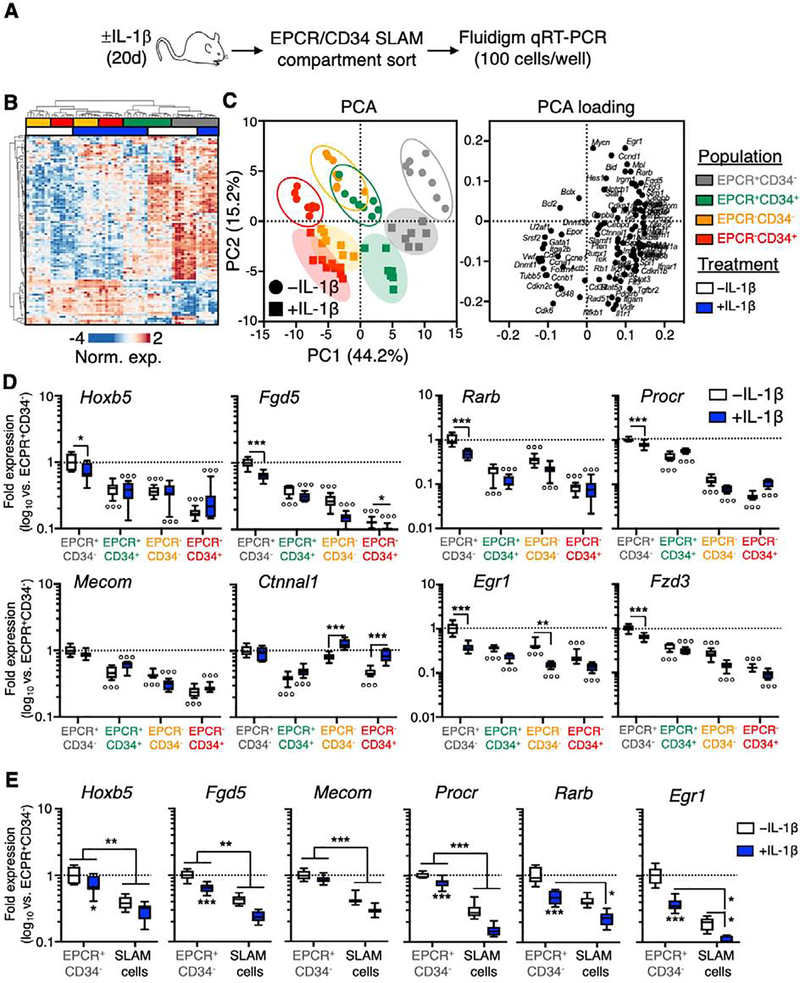
Figure 2. EPCR and CD34 identify molecularly distinct SLAM compartments.
(A) Experimental design of Fluidigm-based molecular analysis of SLAM cells fractionated by EPCR and CD34 expression from mice treated ± IL-1 for 20d. (B) Heatmap and hierarchical clustering analysis, and (C) principal component analysis (PCA; left) and PCA loading plot (right) of Fluidigm gene expression data (N = 7–8 per group). (D) Expression of HSC-associated genes in SLAM cells fractionated by EPCR and CD34 expression, from mice treated ± IL-1 for 20d. (E) Expression of HSC-associated genes in total SLAM cells and EPCR+/CD34− SLAM cells from mice treated ± IL-1 for 20d. Data in (D-E) are expressed as log10 fold expression vs. -IL-1 EPCR+/CD34− SLAM cells. ○ p < 0.05, ○○ p < 0.01, ○○○ p < 0.001 vs. -IL-1 EPCR+/CD34− SLAM cells; * p < 0.05, ** p < 0.01, *** p < 0.001 vs. -IL-1 condition within each fraction, as determined by two-way ANOVA. Data are representative of two independent experiments.