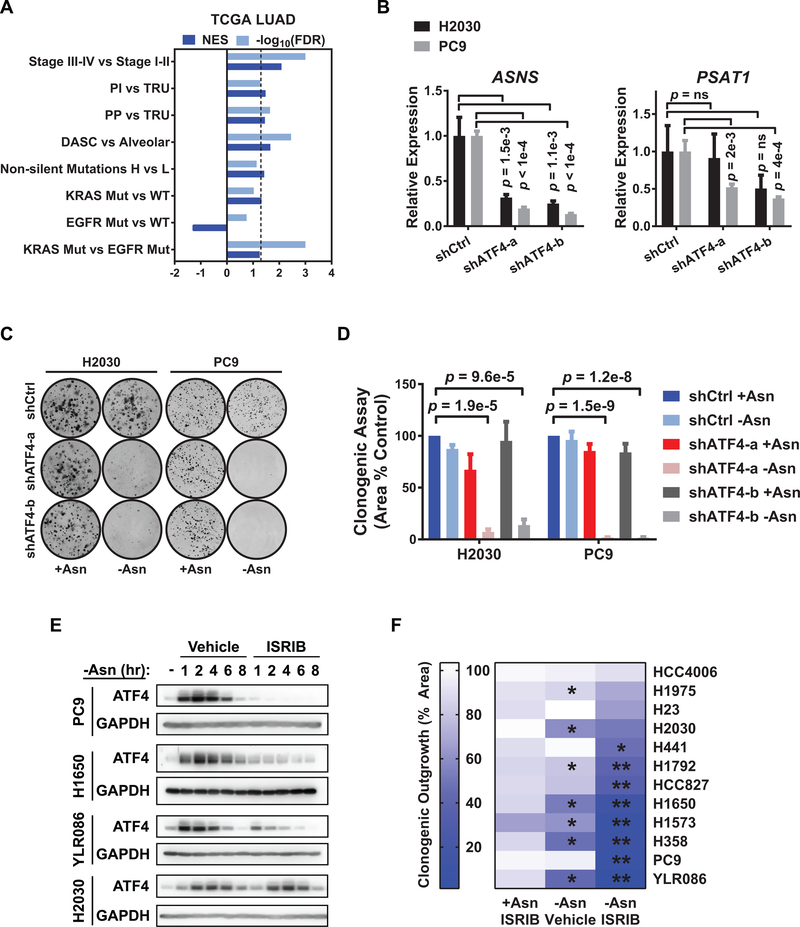
Figure 1. The ISR is activated in diverse molecular subtypes and stages of LUAD and can be inhibited by ISRIB.
A) Gene set enrichment analysis (GSEA) of ATF4 targets in LUAD samples from the TCGA (either n = 489 or n = 275 with paired RNA seq and whole exome sequencing data. Comparisons are stage III-IV (late) versus I-II (early) disease, proximal inflammatory (PI) versus terminal respiratory unit (TRU) tumors, proximal proliferative (PP) versus TRU tumors, DASC-like versus alveolar-like tumors, non-silent mutation high versus low tumors, KRAS mutant versus wildtype tumors, EGFR mutant versus wildtype tumors, and KRAS versus EGFR mutant tumors. NES (normalized enrichment score) and -log10(FDR) are listed for each analysis. Dashed line is FDR < 0.05. B) qRT-PCR of ATF4 target genes involved in amino acid biosynthesis in the H2030 and PC9 cell lines with doxycycline-induced control shRNA or shRNAs against ATF4. Data for two independent shRNAs (a and b) are shown. Relative expression normalized to GUSB with upper and lower limits of a representative experiment shown; n = 2. ns = not significant. C) Representative images of clonogenic assay with the indicated cells plated in +Asn or -Asn media. D) Quantification of C) showing mean and SEM. E) Representative western blots for ATF4 and GAPDH (loading control) from PC9, H1650, YLR086, and H2030 cells following incubation in -Asn media with vehicle (DMSO) or 200 nM ISRIB for the indicated times. F) Heat map depicting quantification of clonogenic assay of a panel of LUAD cell lines treated as indicated. +Asn ISRIB: ISRIB treatment alone. -Asn Vehicle: asparagine deprivation alone. -Asn ISRIB: asparagine deprivation with ISRIB treatment (dual treatment). Asterisks indicate FDR < 0.05 for indicated condition versus (*) +Asn Vehicle or (**) -Asn Vehicle. Unless otherwise noted n = 3 and p-values by Unpaired t-test.