Fig. 3. Comparison of dARC1 and rARC CA-NTD structures.
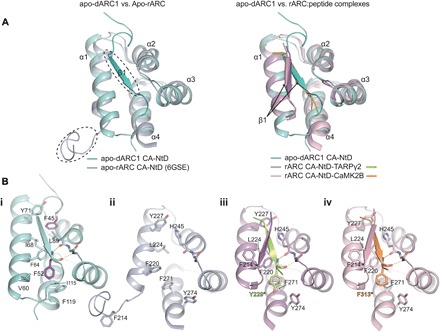
(A) Left: 3D structural alignment of dARC1 CA-NTD (teal cartoon) and apo-rARC CA-NTD (PDB: 6GSE; lilac cartoon). Secondary structure elements are labeled. Circled are the ordered N-terminal β strand of dARC1 and the disordered N-terminal strand of apo-rARC. Right: 3D structural alignment of dARC1 CA-NTD and the peptide-complex structures of rARC CA-NTDs (PDB: 4X3H and 4X3I). The protein backbones are shown in cartoon representation, colored according to the legend. Secondary structure elements are labeled. The arrow indicates the different positioning of the extended N-terminal β strand between the dARC1 and rARC structures. (B, i to iv) Individual views of the structures presented in (A): (i) apo-dARC1, (ii) apo-rARC, (iii) rARC-TARPγ2, and (iv) rARC-CaMK2B. Residues that constitute the hydrophobic NTD cleft are shown in stick format, colored by atom type. In each structure, the side chains of the aromatic residues buried in the interface (F45 and F52, dARC1 CA-NTD; Y229*, rARC CA-NTD–TARPγ2; F313*, rARC CA-NTD–CaMK2B) are colored purple, yellow, and orange, respectively. The conserved main-chain hydrogen bonding interactions between the backbone amide and carbonyl of F52 with the carbonyl of L89 and the amide of Y91 (dARC1), of Y229 with the carbonyl of H245 and the amide of N247 (rARC CA-NTD–TARPγ2), and of F313 (rARC CA-NTD–CaMK2B) with the carbonyl of H245 and the amide of N247 are shown as dashed lines.
