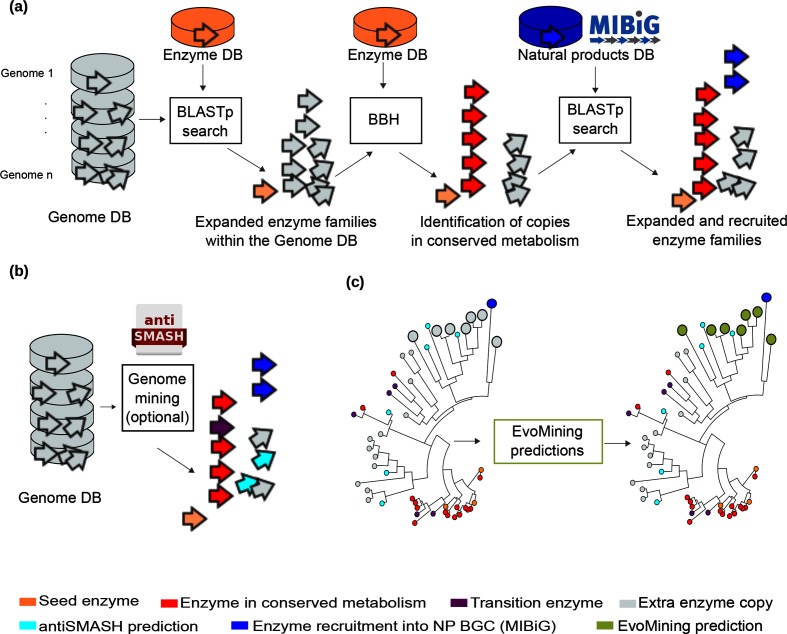
Fig. 1.
EvoMining pipeline, DBs and enzyme’s origin and fate. (a) EvoMining expansion and recruitment pipeline. Homologues and expansions of seed enzymes (orange) from the Enzyme DB are searched by blastp in the Genome DB. The outcome is integrated as the expanded EF. BBH of seed enzymes (red) are marked as conserved metabolism. The EFs are amplified after being compared against the NP DB (blue) to find recruitments defined as enzymes of the family that are part of an MIBiG BGC. (b) Optionally, antiSMASH predictions (cyan) can be added by the user. Enzyme predictions based on antiSMASH that are at the same time marked in red are defined as transition enzymes (purple). (c) EvoMining phylogenetic reconstruction and visualization algorithm. First, a phylogenetic reconstruction of an EF is carried out (left). Extra copies that are neither antiSMASH nor EvoMining predictions are left in grey. Second, on the right-hand side, EvoMining predictions (green), which are those extra copies closer to recruitments (blue) than to conserved metabolic enzymes (red), are shown. These predictions represent novel enzymes devoted to specialized metabolism.