Figure EV3. Structural analysis of the identified 9‐amino acid sequence. (This figure is related to Fig 2 and Appendix Fig S3).
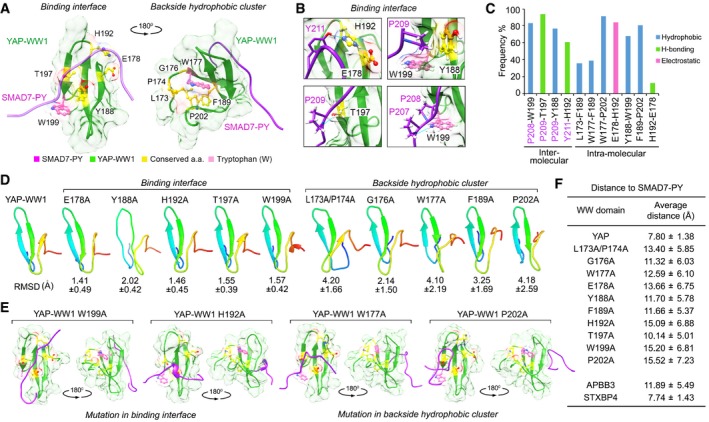
- Illustration of the identified 9‐amino acid residues in the average YAP‐WW1/SMAD7‐PY structure. The initial structure was derived from NMR solution structure (2LTW). SMAD7‐PY peptide was adjusted to 50% transparence to show the residue details on the binding interface.
- Four contact regions within the YAP‐WW1/SMAD7‐PY complex were shown in details from the representative top cluster structures. Residues from SMAD7‐PY motif peptide were labeled in purple. Hydrogen bond is indicated in blue line.
- The binding type and the corresponding frequency rate were shown for the indicated inter‐ and intramolecular residue pairs.
- Simulation analysis of apo YAP‐WW1 domain and its indicated mutants. RMSD value for each mutant simulation (referenced against the average apo YAP‐WW1 domain) was shown.
- Average structures of the indicated YAP‐WW1 mutant/SMAD7‐PY complexes.
- The average distance between SMAD7‐PY motif peptide and the indicated WW domains was summarized in a table.
