Fig. 1.
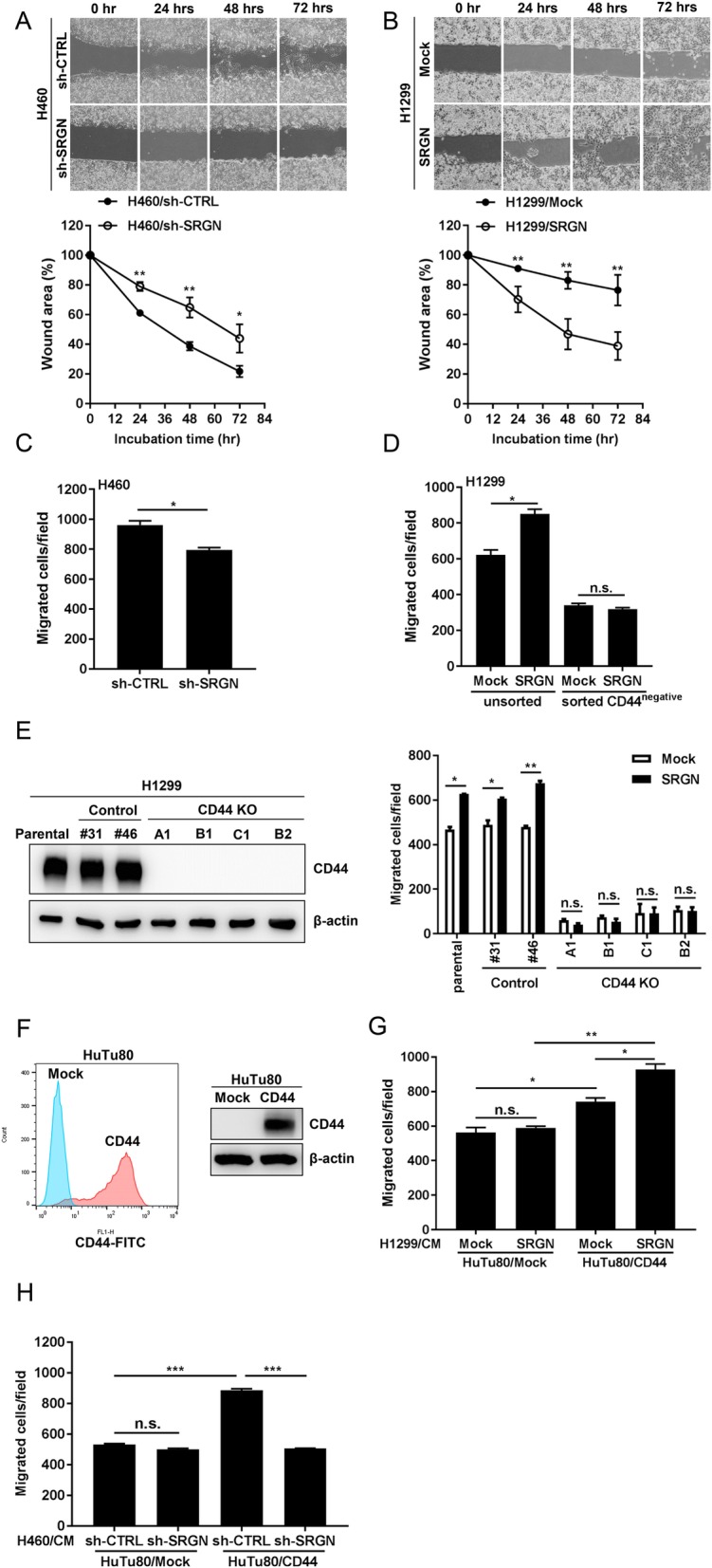
SRGN promotes NSCLC cell migration in a CD44-dependent manner. a-b Wound healing assay was performed by seeding NSCLC-H460 (a) and -H1299 (b) cells in the Culture-Insert 2 Well μ-Plates. After removing the insert, wound closure was determined at 0, 24, 48, and 72 h. Wound area was assessed by ImageJ and normalized with respect to the area at 0 h. c H460/sh-CTRL and H460/sh-SRGN cells were cultured in serum-free medium for 24 h and subjected to Boyden chamber migration assay. Migrated cells were counted in 3 h. d H1299/Mock and H1299/SRGN cells were sorted based on CD44 expression. The unsorted as well as CD44-negative H1299/Mock and H1299/SRGN cells were cultured in serum-free medium for 24 h, and subjected to Boyden chamber migration assay. The migrated cells were counted in 3 h. e The parental H1299 cells and the Control-KO and CD44-KO cell clones generated using the CRISPR/Cas9 system were subjected to Boyden chamber migration assay as described above. The expression of CD44 was shown by western blot. f CD44 expression in HuTu80/Mock and HuTu80/CD44 cells was assessed by flow cytometry (left panel) and western blot (right panel). g-h HuTu80/Mock and HuTu80/CD44 cells were suspended in conditioned media derived from H1299/Mock and H1299/SRGN cells (g) as well as from H460/sh-CTRL and H460/sh-SRGN cells (h), and subjected to Boyden chamber migration assay as described above. The Boyden chamber assays were performed by loading the lower chamber with medium containing 5% FBS in (c) and (h), and 2% FBS in (d), (e) and (g). Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 by Student’s t-test
