Figure 3.
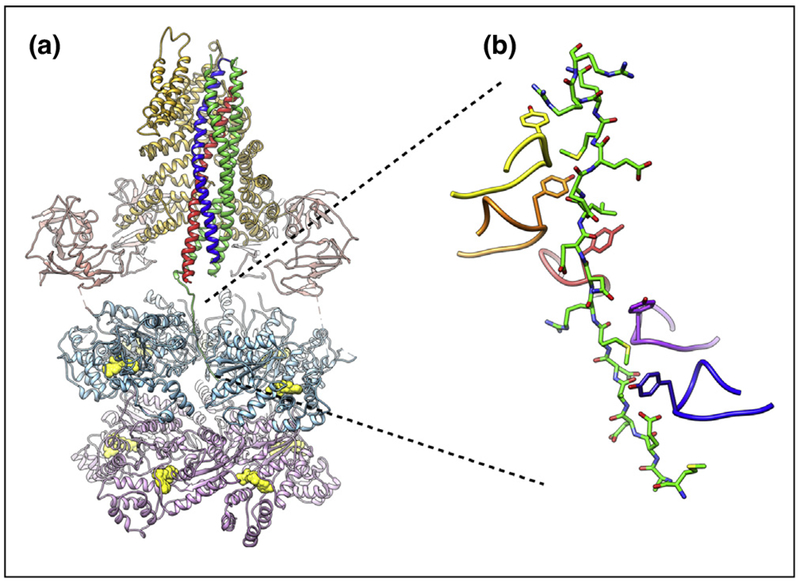
SNARE complex disassembly machinery. (a) EM structure of NSF/αSNAP/SNARE complex at ~3.9 Å resolution [78••] (PDB ID 6MDM). The N, D1, and D2 domains of NSF are colored salmon, light blue, and light purple, respectively. Nucleotides in the D1 and D2 rings are colored yellow. The two αSNAP molecules are shown in gold. The SNARE proteins syntaxin-1a, synaptobrevin-2, and SNAP-25A are colored blue, red, and green, respectively. (b) Close-up view of the interaction between the N-terminal residues of SNAP-25A and the pore of the D1 ATPase ring of NSF. An amino acid essential for SNARE complex disassembly (Y294) is found at the apex of the pore loop of each ATPase subunit; five of six of these tyrosines intercalate between the side chains of the SNAP25 N-terminus, seemingly locking it in place.
