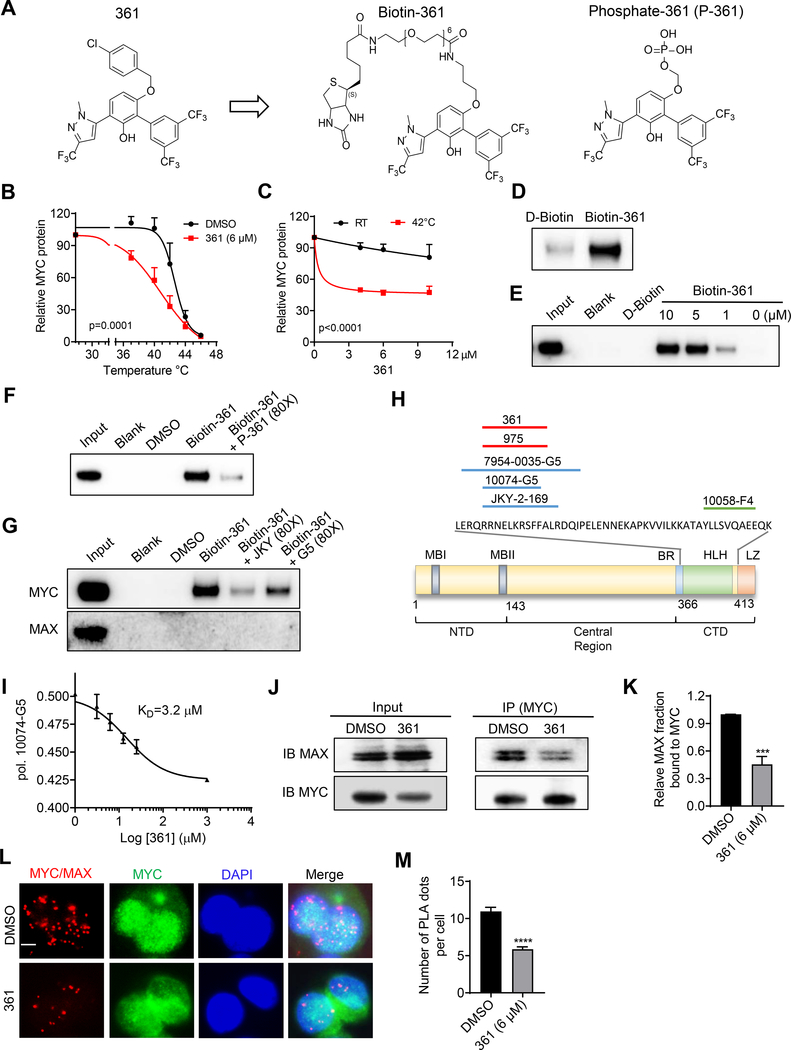
Figure 1.
Identification of MYC Inhibitors
(A) Chemical structures of compound 361, Biotin-361 and Phosphate-361.
(B) Melt curves of MYC protein in cellular thermal shift assay (CETSA) in PC3 cells treated with 361 or DMSO. The graph shows the quantification of MYC protein versus temperature points based on western blot analyses.
(C) 361 CETSA under isothermal condition. Graph shows the quantification of MYC protein at room temperature (RT) 25 °C or 42 °C from cells treated with indicated concentrations of 361.
(D) Western blots for recombinant MYC protein after Biotin-361 (5 μM) or control D-Biotin (5 μM) pulldown.
(E) Western blot analysis on endogenous MYC protein after Biotin-361 pulldown in PC3 cell lysates.
(F and G) Biotin-361 (5 μM) binding to MYC from PC3 cell lysate was analyzed after pre-treatment with Phosphate-361 (F) or compounds G5 or JKY-2–169 (JKY) (G).
(H) Illustration of MYC binding sites of reported MYC inhibitors including G5, JKY, 7594–0035, and F4, as well as 361 and 975 from this study.
(I) 361 binding affinity to MYC was assessed by fluorescence polarization (FP) competition assay. The graph shows 361 at varying concentrations (3.1–25 μM) against G5 (10 μM) binding to MYC353–439 in FP.
(J and K) Western blot showing (J) and quantification of (K) the levels of MAX co-immunoprecipitated with MYC in PC3 cells with or without 1 hr treatment of 361.
(L and M) Representative immunofluorescence (IF) images (L) and quantification (M) of proximity ligation assay (PLA) for MYC/MAX interaction in PC3 cells after 1 hr treatment of 361. Red signals indicate close proximity between MYC and MAX and green fluorescence shows MYC expression at same cell sections (scale bar, 5 μm).
Error bars represent mean ± SEM, n = 3 independent experiments for (B), (C), (I) and (K), n = ~ 200 cells counted/group for (M), and analyzed by two-way ANOVA for (B) and (C), “One site - Fit Ki” analysis and “Binding-competitive” suite for (I), unpaired t-test for (K) and (M) in Prism.***p < 0.001, ****p < 0.0001.
See also Figures S1 and S2