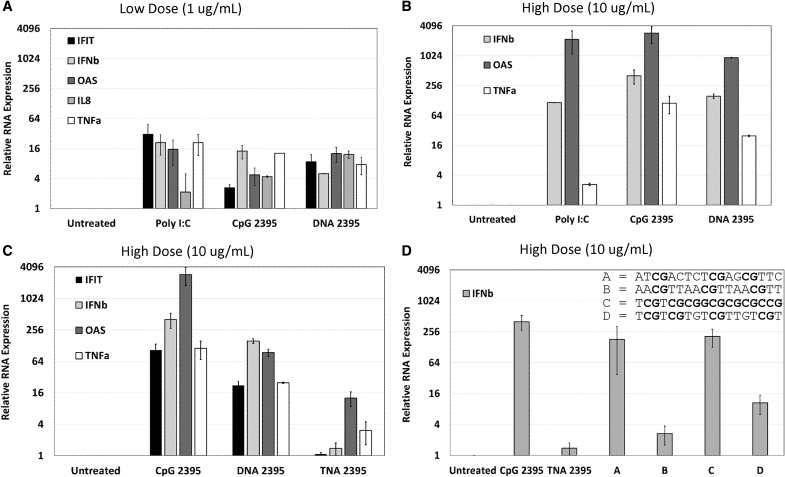
FIG. 1.
Upregulation of mRNA in response to CpG 2395, DNA 2395, TNA 2395, and a TNA panel of variable sequence composition in Ramos cells. For all panels, Ramos cells were plated at a density of 2 × 105 cells in 12-well plates with serum-free medium and stimulated with the indicated ligands (negative control, untreated/vehicle only; positive control, Poly I:C for stimulation of TLR3; test ligands, CpG 2395, DNA 2395, TNA 2395, and Sequences A, B, C, and D) at the indicated concentrations (A, 1 μg/mL; B–D, 10 μg/mL). After 24 h, cells were collected, pelleted, and washed in 1 × PBS. Total cellular RNA was then harvested from each sample. Following DNase treatment of the total cellular RNA, cDNA was synthesized, and quantitative real time-polymerase chain reaction was performed to examine expression of the indicated genes (IFIT, IFNβ, IL-8, OAS, and/or TNFα) and the 18S rRNA housekeeping gene for normalization. Relative quantities were determined using the relative quantity (2−ΔΔCT) method as described in Materials and Methods section. Average quantities are shown for technical replicates measured in triplicate. Error bars represent the standard deviation for three technical replicates within each experiment. A representative experiment is shown in each panel. Experiments were repeated thrice. TNA, threose nucleic acid; CpG, cytosine-phosphate-guanine; PBS, phosphate buffered saline.