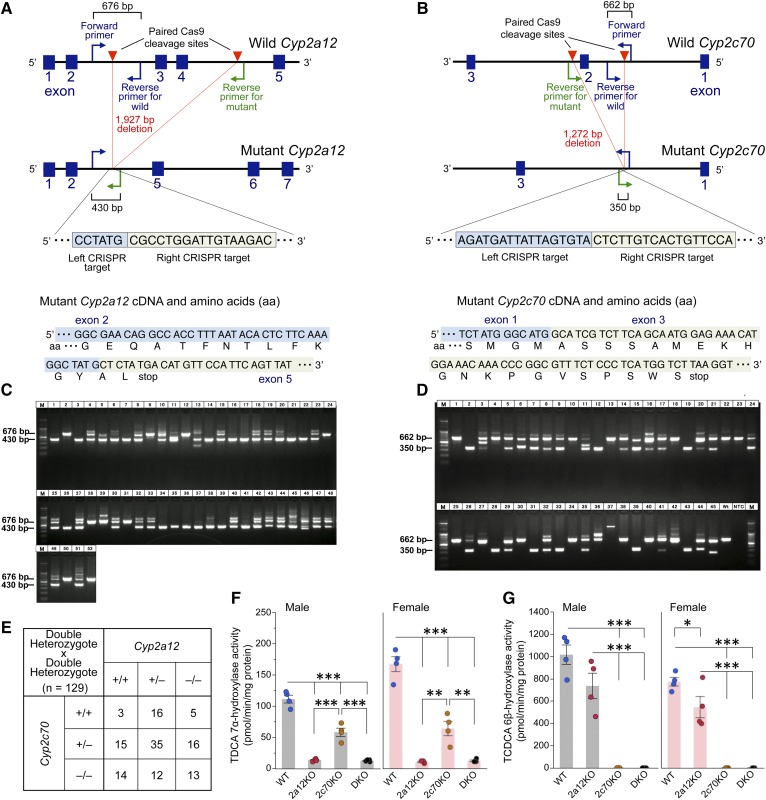
Fig. 2.
Disruption of Cyp2a12 and Cyp2c70 genes markedly reduces hepatic TDCA 7α-hydroxylase and TCDCA 6β-hydroxylase activities. A: CRISPR target site in the Cyp2a12 gene and predicted cDNA and amino acid sequences of mutant Cyp2a12. Primers were designed for genotyping of mice to detect the 1,927 bp deletion of Cyp2a12 induced by Cas9. The forward primer and reverse primer for WT amplify the WT allele (676 bp), and the forward primer and reverse primer for mutant amplify the mutant allele (430 bp). B: CRISPR target site in the Cyp2c70 gene and predicted cDNA and amino acid sequences of mutant Cyp2c70. Primers were designed for genotyping of mice to detect the 1,272 bp deletion of Cyp2c70 induced by Cas9. The forward primer and reverse primer for WT amplify the WT allele (662 bp), and the forward primer and reverse primer for mutant amplify the mutant allele (350 bp). C: PCR genotyping of Cyp2a12−/− (430 bp), Cyp2a12+/− (430 bp and 676 bp), and Cyp2a12+/+ (676 bp) in pups (n = 52) for the screening of F0 mice. D: PCR genotyping of Cyp2c70−/− (350 bp), Cyp2c70+/− (350 bp and 662 bp), and Cyp2c70+/+ (662 bp) in pups (n = 45) for the screening of F0 mice. E: Genotypes of mice (n = 129) obtained by crossing double heterozygous Cyp2a12+/−Cyp2c70+/− animals. F0 mice were bred with WT C57BL/6J mice to obtain F1 heterozygous Cyp2a12+/− and Cyp2c70+/− mice, and these were crossed to produce Cyp2a12+/−Cyp2c70+/− mice. Then, these double heterozygous mice were crossed to obtain Cyp2a12−/−Cyp2c70+/+ (2a12KO), Cyp2a12+/+Cyp2c70−/− (2c70KO), and Cyp2a12−/−Cyp2c70−/− (DKO) mice. F, G: Hepatic TDCA 7α-hydroxylase (F) and TCDCA 6β-hydroxylase (G) activities in WT, 2a12KO, 2c70KO, and DKO mice (n = 4). Each column and error bar represents the mean and SEM. *P < 0.05, **P < 0.005, and ***P < 0.0001 were considered significantly different by the Tukey-Kramer test.