Figure 1. Strategy for designing metabolite sensors using Corn.
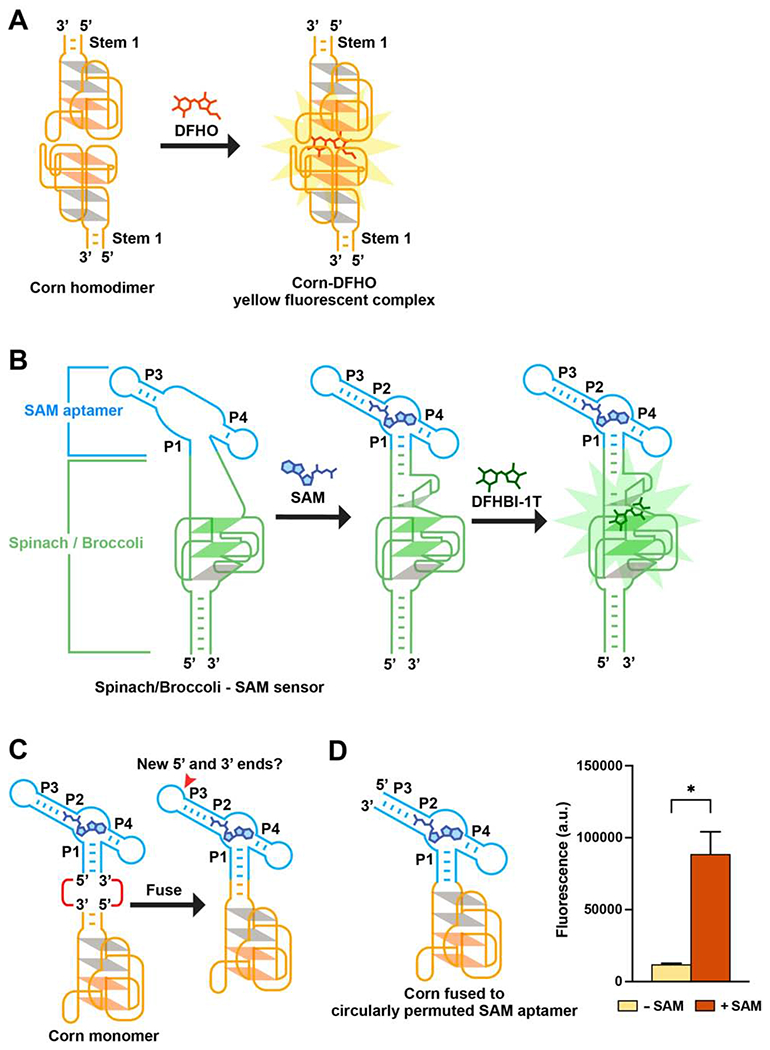
(A) Corn contains one helical stem (Stem 1) unlike Spinach or Broccoli. DFHO binds the Corn dimer interface, between the apical G-quadruplex in each Corn monomer. Orange parallelograms indicate the two G-quartets. Grey parallelograms indicate two mixed-sequence tetrads.
(B) Schematic diagram of the Spinach-based SAM sensor. SAM induces folding of P1, which serves as a transducer domain. P1 folding enables Spinach folding and binding to DFHBI-1T (right).
(C) Since the P1 stem has the 5’ and 3’ ends of the SAM aptamer, fusing Corn to the SAM aptamer P1 stem would result in an RNA lacking 5’ and 3’ ends. The P3 stem contains the new 5’ and 3’ ends.
(D) Corn fused to the circularly permuted SAM aptamer exhibits SAM-induced fluorescence. In vitro transcribed RNA (1 μM) was incubated with 0.1 mM SAM or water for 1 h (37°0 and 1 mM MgCl2). Addition of SAM resulted in a 7.4-fold increase in fluorescence (ex 505 nm; em 545 nm). Mean and SEM values are shown (n = 3). * P = 0.0384
