Abstract
Background and Aims
Heterogeneity in Crohn’s disease [CD] provides a challenge for the development of effective therapies. Our goal was to define a unique molecular signature for severe, refractory CD to enable precision therapy approaches to disease treatment and to facilitate earlier intervention in complicated disease.
Methods
We analysed clinical metadata, genetics, and transcriptomics from uninvolved ileal tissue from CD patients who underwent a single small bowel resection. We determined transcriptional risk scores, cellular signatures, and mechanistic pathways that define patient subsets in refractory CD.
Results
Within refractory CD, we found three CD patient subgroups [CD1, CD2, and CD3]. Compared with CD1, CD3 was enriched for subjects with increased disease recurrence after first surgery [OR = 6.78, p = 0.04], enhanced occurrence of second surgery [OR = 5.07, p = 0.016], and presence of perianal CD [OR = 3.61, p = 0.036]. The proportion of patients with recurrence-free survival was smaller in CD3 than in CD1 (p = 0.02, median survival time [months] in CD1 = 10 and CD3 = 6). Overlaying differential gene expression between CD1 and CD3 on CD subgroup-associated genetic polymorphisms identified 174 genes representing both genetic and biological differences between the CD subgroups. Pathway analyses using this unique gene signature indicated eukaryotic initiation factor 2 [eIF2] and cyclic adenosine monophosphate [cAMP] signalling to be dominant pathways associated with CD3. Furthermore, the severe, refractory subset, CD3, was associated with a higher transcriptional risk score and enriched with eosinophil and natural killer T [NKT] cell gene signatures.
Conclusion
We characterized a subset of severe, refractory CD patients who may need more aggressive treatment after first resection and who are likely to benefit from targeted therapy based on their genotype and tissue gene expression signature.
1. Introduction
Inflammatory bowel disease [IBD] comprises a variety of disorders associated with chronic inflammation of the gastrointestinal tract. Classically, IBD has been assigned as either ulcerative colitis [UC] or Crohn’s disease [CD]. Crohn’s disease most commonly affects the small bowel [SB] and exhibits a diversity of clinical phenotypes, including stricturing, internal penetrating, and perianal CD [pCD]. This heterogeneity provides a challenge for the development of effective therapies and may be one of the reasons behind drug development failures and limited efficacy with existing therapies, including the anti-TNF agents.1,2
Therefore, defining a unique signature associated with the various clinical phenotypes of CD would better identify homogenous CD patient subgroups with similar pathways and mechanistically distinct disease subtypes.3 These defined patient subgroups may help us to arrive at better understanding of the underlying pathology, provide patient stratification, aid in selection from existing therapies, and ultimately help in development of personalized or precision approaches to effectively treat IBD patients.4–6
There have been a number of recent attempts to use transcriptomics to classify CD subtypes.7–9 Both adult and paediatric CD patients have been classified in clinically distinct subgroups associated with either colon-like or ileum-like gene expression profiles.8 Transcriptional profiling of T cells from IBD patients has revealed subgroups with varying disease course.7 However, inclusion of the genetic contribution to disease prognosis,10 along with susceptibility to IBD, in studies such as these would provide a mechanism for both linking a transcriptional risk profile and identifying a potential therapeutic target. Transcriptional risk scores [TRSs] calculated in a paediatric CD cohort have connected known IBD genetic variants to expression quantitative trait loci [eQTL].9 Transcriptional risk scores could identify patients who would progress to complicated disease over time.
In this work, we utilized a cohort of CD patients who had undergone SB resection as part of their treatment, to dissect the CD-related pathogenic heterogeneity that existed in a group of refractory patients with varying disease course. We focused on identifying clinically relevant subgroups, using both expression and genetic data from uninvolved ileal tissue taken from SB resections. Within this heterogeneous, refractory CD population, we identified clinically distinct patient subgroups with varying disease severity. We then overlapped the genetic- and transcriptomics-based signals from the same patients to define molecular signatures that may help in the development of personalized therapies.
2. Materials and Methods
2.1. Sample cohorts
Transcriptomic data was generated on SB tissue as previously described.11 Briefly, uninvolved tissue from formalin-fixed paraffin-embedded [FFPE] SB resection margins of subjects requiring surgery at Cedars-Sinai Medical Center for Crohn’s disease was identified. Whole-thickness ileal tissue was scraped from the FFPE tissue sections, followed by RNA extraction using an RNeasy FFPE kit [Qiagen] according to the manufacturer’s instructions. A Transplex Whole Transcriptome Amplification kit [WTA2; Sigma] was used for cDNA synthesis and amplification. Subsequent purification of the cDNA product was performed with a PCR Purification kit [Qiagen]. Sample quality was confirmed using an Agilent Bioanalyzer. We used the same methods for sample selection as reported previously by VanDussen KL et al.12 Instead of using RNA integrity number [RIN] scores as selection criteria, samples were selected for study only if >20% of the RNA fragments were 200 base pairs or greater in length, as determined with the Agilent Bioanalyzer software. This ensured that only the most intact FFPE samples were included, and any samples with a high level of degradation [with only short fragments] were excluded. For samples passing quality control, Cy5 labelling with the ULST Fluorescent Labeling kit [Kreatech] and hybridization [performed in duplicate for each sample] to Whole Human Genome 4x44k Microarrays [Agilent] was performed.
2.2. Expression data processing and clustering
Single-channel microarray expression data extracted using Agilent feature extraction software was received from the Genome Technology Access Center at Washington University in St Louis. Raw expression data available in technical duplicates was normalized using the LIMMA package implemented in R version 3.2.1.13 All the gene expression data, including the sample metadata, the Agilent raw data, and the processed data for all of the 157 samples can be accessed at Gene Expression Omnibus using accession number GSE120782. The expression data preprocessing included background correction of the expression data, followed by log2-transformation and quantile-normalization. Unsupervised hierarchical clustering of expression data was used to remove outlier subjects.
Differential gene expression analysis was done by class comparison in BRB array tools using probe gene expression corresponding to each of the three subgroups. Sometimes, a gene filter cut-off was applied during class comparison, in which case a gene was excluded if <20% of the expression data had at least a 1.5-fold change in either direction from the gene’s median value.
The transcriptomic data was generated in two batches [n = 100, n = 57] and was analysed separately as well as in a merged dataset. Only Caucasian subjects [≥75% as defined by Admixture14] were retained in analyses. Sample outliers in each expression cohort were removed if the technical duplicates did not cluster. A total of 139 Caucasian CD patients were included in this study. Hierarchical and kmeans clustering [implemented in R] using the first three principal components [PCs] in normalized expression data with SB85 indicated the presence of three sample clusters [CD1, CD2, and CD3]. Combining SB85 with SB54 after removal of batch effects preserved these three subgroups. We used the ‘removebatcheffect’ function in the LIMMA R package13 to remove the batch effects between the SB85 and SB54 datasets.
We validated the presence of three patient subgroups in our combined expression data using a non-heuristic, model-based clustering method. This was implemented using the ‘mclust’ R package, which is based on using Gaussian finite mixture models.15 With this method, the data is assumed to be part of a distribution that is a mixture of two or more subgroups, and each group is modelled by a Gaussian distribution with a specific mean vector, covariance matrix, and associated probability in the mixture. The advantage of using model-based clustering is the recommendation of the number of clusters/subgroups present and of the best model to fit the data. Using the Bayesian information criterion score, an optimal number of three subgroups was recommended by application of the mclust package on the merged dataset [supplementary Figure S2].
2.3. Clinical phenotyping and genotyping
Clinical data, including patients’ gender, age at diagnosis, disease location and behaviour [according to the Montreal Classification], and surgical history were collected as previously described.16
Genotyping was performed at the Cedars-Sinai Medical Center using the Illumina Immuno-BeadChip array as previously described.2,16,17 Markers were excluded from analysis based on: Hardy–Weinberg Equilibrium p ≤ 0.001; genotyping rate <98%; and minor allele frequency <5%. Related individuals [Pi-hat scores > 0.25] were identified using identity-by-descent and excluded from analysis using PLINK.18 Admixture was used to generate ethnicity proportion estimations for all individuals.14 Only subjects identified by admixture as Caucasian [proportion ≥ 0.75] were included in the analysis; thus, a total of 139 independent Caucasian samples were retained in the analysis. Principal component analysis [PCA] was performed using Eigenstrat, and the top two PCs were included as covariates in the analysis to adjust for potential population substructure.19 We performed genetic associations [logistic regression with PC adjustment] for the presence or absence of a given subgroup [CD1/CD2/CD3], using genotype data for the 139 subjects in the combined cohort.
2.4. Overlap of genetic loci and differentially expressed genes underlying the subgroups
We first compiled a list of genes corresponding to genetic loci [p < 0.05] that were associated uniquely with either the CD1 subgroup or CD3 subgroup. We excluded any shared genes corresponding to genetic associations between the CD1 and CD3 subgroups because we wanted to locate genetic loci unique to each of the subgroups. We then overlapped this list of genes based on genetic associations with the differential expression [DE] gene list based on 4380 gene expression probes. This gave us a list of 174 genes [supplementary Table S2] with unique genetic association with either the CD1 or CD3 subgroups and also the DE genes between the two subgroups.
2.5. eQTL mapping
eQTL mapping was implemented in the Matrix eQTL R package using the available expression and genotype data for n = 26 [CD1] and n = 25 [CD3] independent Caucasian samples.20 We also generated eQTLs considering all the 139 samples together as part of determining eGenes to calculate TRSs. Associations between genotype and probe expression level were performed using a linear regression model with additive genotype effects. All associations were adjusted for gender and population substructure using the first two PCs of genetic data. Gene bounds were defined using a 1 Mb window around the transcription start position of a given gene as obtained from the UCSC Genome Browser. For cis-eQTL mapping, a 1 Mb cis distance from gene bounds was used. Cis-eQTLs were defined as association signals from single nucleotide polymorphisms [SNPs] located within 1 Mb of each of the gene bounds. False discovery rates [FDRs] were estimated to correct for multiple testing using Matrix eQTL according to the Benjamini and Hochberg method.
2.6. TRS calculation
We used the methods described in Marigorta et al.9 to calculate the TRS. Of the 232 known IBD loci, 122 are either cis-eQTLs or in strong linkage disequilibrium [LD] [r2 > 0.8], with at least one cis-eQTL in peripheral blood. This corresponds to a total of 163 [157 unique] corresponding eGenes, i.e. ~1.3 candidate genes per SNP. We determined 139/157 eGenes to be present [with a nominal p-value < 0.05] in the cis-eQTL dataset of all the 139 samples. All 139 eGenes had cis-eQTLs in known regions [as defined by Jostins et al. or Liu et al.21,22] in the SB139 cis-eQTL dataset. Transcript abundance in the SB139 cohort for the short-listed 139 eGenes was standardized and polarized according to direction of risk, as noted previously.9,17,22 Transcript abundance in cases where low expression was associated with risk were flipped. Summation over all eGenes gave the TRS, which was further standardized.
2.7. Cell-type-specific enrichment analysis using xCell
We used xCell23 to generate cell-type-specific signatures associated with the three subgroups. The entire gene expression set corresponding to SB139 cohort was the input for the enrichment analysis using xCell. The most differential cell-type-specific enrichment scores across samples were examined for statistical significance for the three subgroups.
2.8. Eosinophil count using H&E staining
We randomly chose FFPE slides for 67 out of the 139 patients [CD1 = 18, CD2 = 27, and CD3 = 22] and stained them with H&E stain. Slides were scanned at ×20, and eosinophils were manually counted by a trained pathologist in six [300 × 300 μ] random fields of the lamina propria, with areas with outliers or artifacts excluded.
2.9. Pathway analysis
Pathway analysis was accomplished through the use of Qiagen’s Ingenuity® Pathway Analysis [IPA®, Qiagen, Redwood City, www.qiagen.com/ingenuity]. Pathway analysis using the set of DE genes separating the subgroups was performed in IPA, along with a diseases and biological function analysis. Class comparison analysis in BRB array tools with the gene-filter criterion turned on gave a list of 4380 DE gene expression probes, but with the filter turned off gave a much larger list of DE gene expression probes [>18 000 probes] between the subgroups.
2.10. Study approval
Tissue samples and genetic data were obtained by the Material and Information Resources for Inflammatory and Digestive Diseases [MIRIAD] IBD Biobank after the patients’ informed consent and approval by the IRB of the Cedars-Sinai Medical Center [protocol #3358].
2.11. Statistical analysis
All the statistical analyses were performed using R 3.2.1.24
3. Results
3.1. Presence of three subgroups in small bowel resection expression data
We analysed transcriptomic data generated using uninvolved ileal tissue from CD patients who underwent SB resection using methods previously described.11 The data was generated in two batches [n = 100, n = 57] and analysed both separately and as a merged dataset.
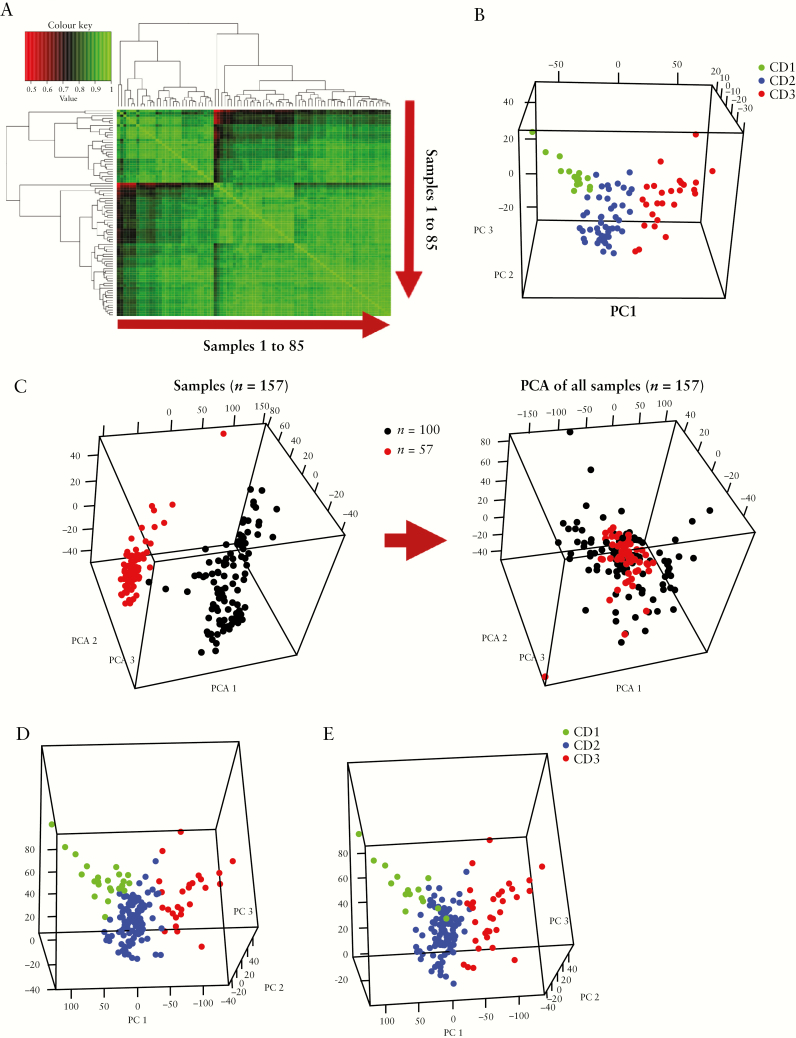
We first looked at sample correlation of normalized, background-corrected expression data after removing outliers and non-Caucasian samples from our larger cohort of 100 samples. We called this cohort SB85 [post–quality control, n = 85]. Figure 1A shows the heat map of the Pearson correlation coefficient between the samples using normalized expression probe data in the SB85 cohort. The heat map revealed the presence of three patient subgroups. We reduced the dimensionality of the expression dataset using PCA and the top three PCs [which explained most of the variance in the expression data] also indicated that the samples clustered into three subgroups. Multiple clustering methods [hierarchical, k-means clustering, and model-based clustering; see Methods] were applied to allocate samples to each subgroup. Figure 1B shows the PCA plot for the SB85 cohort, highlighting the three CD patient subgroups, CD1, CD2, and CD3. The three subgroups in SB85 were homogenous in terms of Jewish ethnicity and disease behaviour [supplementary Figure S1].
Figure 1.
The presence of three patient subgroups in small bowel resection mRNA expression. [A] A heatmap showing the Pearson correlation coefficient between samples using normalized probe gene expression in the SB85 cohort. [B] Three sample clusters in the principal component analysis plot using the top three principal components in the expression data in the SB85 cohort. The sample clusters CD1, CD2, and CD3 were detected using hierarchical and kmeans clustering. [C] The batch effect present in the two cohorts [left] was removed [right], and the cohorts were merged to increase power for genetic and clinical phenotype associations. [D, E] The presence of the three CD patient subgroups was confirmed in an expanded cohort [SB139] using kmeans/hierarchical clustering [left] and a Gaussian model–based clustering method [right].
We then sought to increase the power of our study by adding additional subjects from the SB54 cohort. We therefore merged the two expression cohorts into a combined cohort to increase the sample size to perform further associations, because we were underpowered to draw conclusive results using only the SB54 dataset. Batch effects in the expression data were removed, and the two datasets were merged [combined cohort SB139; Figure 1C]. Both the SB85 and SB54 cohorts were similar in terms of baseline characteristics. There were no statistically significant differences in average age at diagnosis [24 years for both cohorts], gender, disease location and behavior, pCD occurrence, or time to second surgery or follow-up between the two cohorts.
The PCA plot of our combined cohort SB139 confirmed the presence of three distinct patient subgroups [Figure 1D] using kmeans/hierarchical clustering. The CD1 and CD3 subgroups were found to be the most distant as seen in the 3-D PCA plot [Figure 1B–D]. We validated the presence of three patient subgroups in our combined expression data using a non-heuristic, model-based clustering method [see Methods]. A comparison of the three subgroups from model-based clustering and kmeans/hierarchical clustering indicated mostly similar sorting of samples into the three subgroups [Figure 1D and E].
3.2. Clinical variables associated with the patient subgroups indicated patients in the CD3 cluster to be more severely affected
Phenotypic differences in the transcriptomic-based patient subgroups and clinical variables associated with the subjects were investigated. Table 1 shows baseline clinical characteristics of each of the three patient subgroups in our combined cohort [Total, n = 139; CD1, n = 26; CD2, n = 88 and CD3, n = 25].
Table 1.
Baseline characteristics of patients in the three subgroups of SB139 cohort
| CD1 [n = 26] | CD2 [n = 88] | CD3 [n = 25] | |
|---|---|---|---|
| Age at diagnosis, year ± SD | 21.89 ± 12.99 | 25.38 ± 13.61 | 22.94 ± 11.02 |
| Gender [Female], n [%] | 12 [46.1] | 38 [43.1] | 20 [80] |
| Disease location, n [%] | |||
| L1, ileum | 14 [53.1] | 39 [44.3] | 9 [36] |
| L2, colon | 0 [0] | 2 [2.3] | 0 [0] |
| L3, ileocolon | 12 [46.1] | 44 [50] | 16 [64] |
| L4, upper GI | 4 [15.3] | 7 [7.9] | 3 [12] |
| Disease behavior, n [%] | |||
| B1, non-stricturing non-penetrating | 1 [3.8] | 2 [2.2] | 2 [8] |
| B2, stricturing | 11 [42.3] | 38 [43.1] | 13 [52] |
| B3, penetrating | 14 [53.8] | 48 [54.5] | 10[40] |
| Perianal disease, n [%] | 6 [27.7] | 24 [27.2] | 13 [52] |
| Second surgery, n [%] | 4 [15.4] | 24 [27.2] | 12 [48] |
| Recurrence, n [%] | 12 [46] | 23 [26] | 19 [76] |
n = number of positive occurrences of phenotype.
We observed clinical differences between the most distant subgroups [CD1 and CD3] of SB139. CD3 was associated with higher occurrence of second surgery [OR = 5.07, p = 0.016] and presence of pCD [OR = 3.61, p = 0.036] [Table 2]. Compared with CD1, CD3 was enriched for subjects with increased disease recurrence after first surgery [OR = 6.78, p = 0.04]. No significant differences were found when comparing all three subgroups simultaneously for differences in various clinical phenotypes, including disease location, CD disease behaviour information based on Montreal classification,16,25 [described as B1, non-stricturing, non-penetrating; B2, stricturing; and B3, internal penetrating diseases] and occurrence of second surgery. Gender was also associated with clustering [Table 2], with the more severely affected CD3 subgroup consisting of a higher percentage of females. Given our small sample size, in separate multivariate models with gender as a covariate, the significance of the association of clustering [CD1 and CD3] with pCD was reduced [OR = 2.82, p = 0.1], but with occurrence of second surgery remained significant [OR = 4.62, p = 0.03].
Table 2.
The most distant subgroups, CD1 and CD3, in the merged SB139 cohort are clinically different. The CD3 cluster was associated with increased second surgeries and higher pCD occurrence. Gender was also associated with clustering
| [CD1, n = 26] | [CD3, n = 25] | |||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype | No | Yes | % Yes | No | Yes | % Yes | OR [95% CI] | p |
| Second surgery | 22 | 4 | 15.38 | 13 | 12 | 48.00 | 5.07 [1.44–21.31] | 0.016 |
| Perianal disease | 20 | 6 | 27.27 | 12 | 13 | 52.00 | 3.61 [1.12–12.75] | 0.036 |
| Male | Female | % Female | Male | Female | % Female | OR [95% CI] | p | |
| Gender | 14 | 12 | 46.15 | 5 | 20 | 80.00 | 4.66[1.41–17.59] | 0.015 |
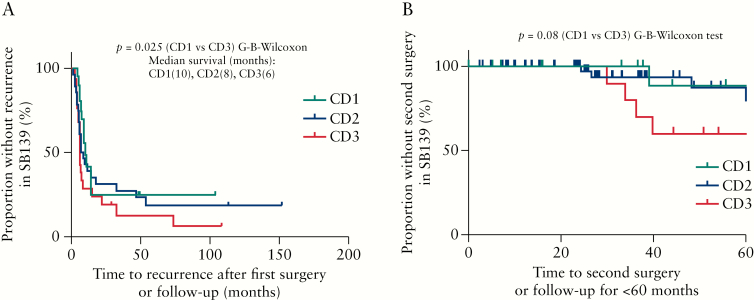
Survival analysis using time from first surgery to recurrence or last follow-up indicated that the time to recurrence in CD3 was shorter than in CD1 [p = 0.02] [Figure 2A]. The median time to recurrence from first surgery [in months] for CD1 was 10, for CD2 was 8, and for CD3 was shortest at 6 months [Figure 2A]. Using time from first to second surgery or last follow-up within 5 years suggested a greater proportion of patients did not have second surgery in CD1 compared with those in CD3 [p = 0.08] [Figure 2B]. These data suggest that the CD3 cluster contained individuals with a more severe disease course.
Figure 2.
The CD3 patient subgroup had faster disease recurrence and there was a trend towards a shorter time to second surgery compared with the CD1 subgroup. [A] The survival analysis [Gehan–Breslow–Wilcoxon test] using time from first surgery to recurrence or last follow-up indicated that the proportion of recurrence-free survival in the CD3 cluster was smaller than in the CD1 subgroup [p = 0.02, median survival time [months], CD1 = 10, CD2 = 8, and CD3 = 6. [B] Using time from first to second surgery or last follow-up within 5 years indicated a greater proportion of patients being without a second surgery in CD1 compared with those in CD3 [marginally significant, p = 0.08] in the combined cohort of SB139.
3.3. Differential gene expression across the subgroups revealed specific expression signatures associated with CD3 compared with the less severely affected CD1 subgroup
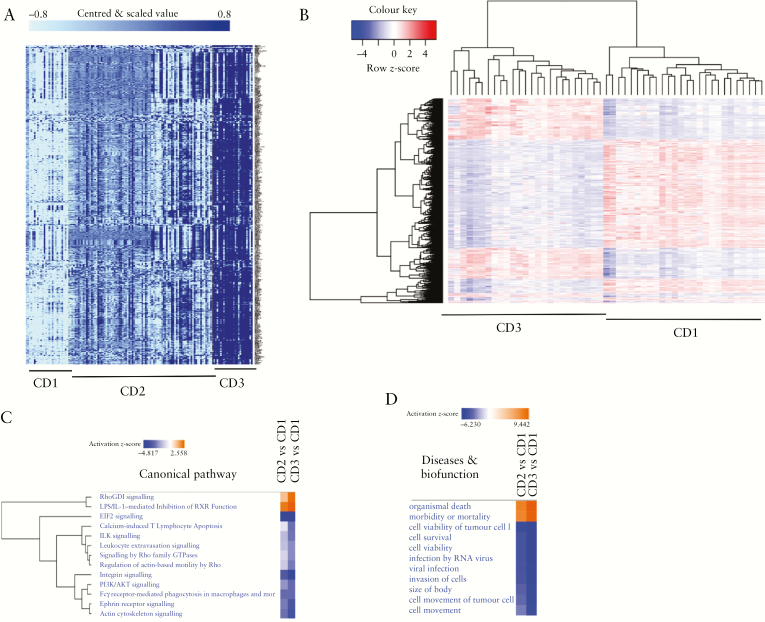
We performed a class comparison, using gene expression corresponding to the three subgroups, on the SB139 dataset. This indicated a list of 4380 gene expression probes as being significantly different [FDR < 0.001] between each of the pairs from the three subgroups. Figure 3A shows the heat map of the DE genes in the three subgroups. Figure 3B shows the DE genes between CD1 and CD3 subgroups, with the dendrogram above the column showing clustering of the CD1 and CD3 samples together.
Figure 3.
Genes differentially expressed between the patient subgroups. [A] Class comparison of the three groups using probe-expression data of SB139 performed in BRB array tools indicated 4380 gene expression probes to be significant [false discovery rate < 0.001]. A gene filter cut-off was used where a gene was excluded if <20% of expression data had at least a 1.5-fold change in either direction from the gene’s median value. The darker-blue shade shows higher expression. [B] Heat map of genes differentially expressed between CD1 and CD3 patient subgroups with expression row-scaled across samples using class comparison results in A. The dendrogram above the columns indicated clustering of samples in CD1 and CD3 subgroups. [C] Comparison of pathways and [D] disease and biofunction in IPA. Activated [orange] and downregulated [blue] in either the CD2 or CD3 subgroups with respect to the CD1 subgroup. EIF2, actin, and integrin signalling was down in CD3 compared with in CD1, and organismal death was activated in CD3 compared with in CD1.
Pathway analysis identified eukaryotic initiation factor 2 [eIF2], actin cytoskeleton, and integrin signalling to be downregulated in CD3 versus CD1, as indicated by negative activation z-scores [Figure 3C], while organismal death was activated in CD3 versus CD1 subgroups [Figure 3D]. Pathway analyses using an expanded list of >18 000 DE gene expression probe sets [see Methods and supplementary Table S1] also indicated that eIF2 signalling was downregulated in CD3 versus CD1 subgroups, while RhoGD1 signalling was activated in CD3 compared with CD1 [supplementary Figure S3].
3.4. Overlap of the genetic and gene expression signatures defining the CD3 subgroup revealed specific pathways to be driving the CD3 phenotype
We determined whether there were differences in the genetic loci underlying susceptibility and associated disease severity for the three CD subgroups identified via transcriptomics [see Methods and supplementary Figure S4].
Having determined the genetic as well as transcriptomic loci for the subgroups in the SB139 merged dataset, we focused on candidate genes that appeared in our genetic associations as well as in the DE gene list. We found that 174 genes associated specifically with either CD1 or CD3 were also differentially expressed between the two subgroups [see Methods and supplementary Table S2].
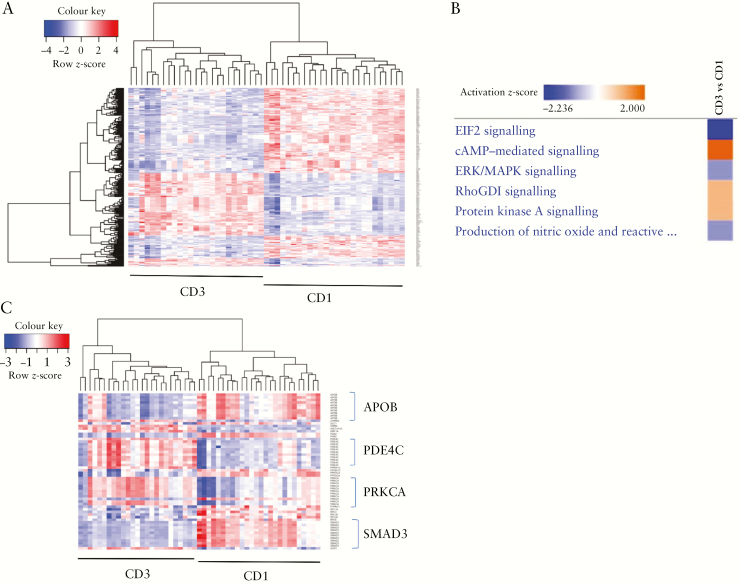
A heat map showing the differential expression of this overlapping list of 174 genes is depicted in Figure 4A. Pathway analyses using these genes and the associated expression fold-changes between CD3 versus CD1 subgroups indicated eIF2 signalling was downregulated in the CD3 subgroup compared with the CD1 subgroup, and cyclic adenosine monophosphate [Camp]-mediated signalling was activated in CD3 compared with CD1 [Figure 4B]. Figure 4C shows a heat map highlighting some key genes [APOB, PDE4C, PRKCA, and SMAD3] involved in pathways differentially regulated between the CD3 and CD1 subgroups. Thus, we identified key genes based on genotype and/or differential expression that differentiated the clinically distinct CD1 and CD3 patient subgroups [Table 3] in the combined SB139 cohort.
Figure 4.
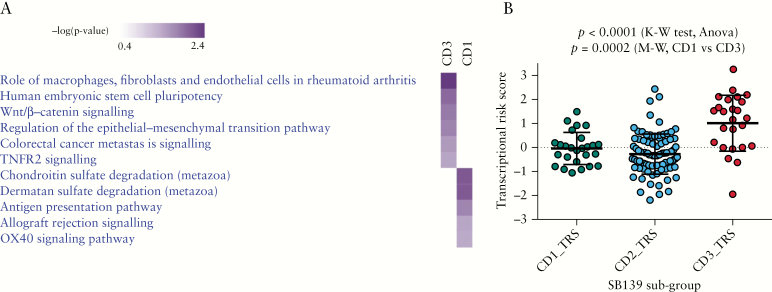
Overlap of genetic loci [genes] associated uniquely with either CD1 or CD3 and differentially expressed genes [CD3 vs CD1] reveal specific pathways driving the CD3 phenotype. [A] Heat map focusing on overlapping genes [n = 174] with exclusive genetic association with either CD1 or CD3 subgroup and also differentially expressed between the two subgroups. [B] Pathways defining the CD3 subgroup compared with the CD1 subgroup, using 174 genes defined above. [C] Heat map highlighting some key genes involved in the CD3 vs CD1 pathways mentioned in B.
Table 3.
Key genes differentiating the clinically distinct CD3 and CD1 patient subgroups based on genotype and expression in the SB139 cohort. The molecular signatures of these patient subgroups in severely affected CD populations can help in development of personalized therapies
| Gene | CD3 vs CD1 fold change | Genetic association [p < 0.05] |
|---|---|---|
| PDE4C | 2.12 | Associations with CD3 |
| ICAM3 | 2.41 | Associations with CD3 |
| SMAD3 | –2.41 | Association with CD1 |
| IL18BP | 1.48 | Association with CD3 |
| DAPK1 | NA | Associations with CD3 |
| SHANK3 | –2.59 | NA |
| OSMR | 1.95 | NA |
3.5. eQTL analyses revealed differences in the CD1 and CD3 subgroups
Using the genetic and transcriptomic data for the SB139 cohort, we performed eQTL analyses to determine genetic loci that directly regulated local gene expression in the CD subgroups of varying severity, with CD1 being less severely affected and CD3 being more severely affected. Cis-eQTL analysis revealed that the CD1 and CD3 subgroups have mostly distinct signatures. All the cis-eQTLs with FDR < 0.001 were unique to either the CD1 or CD3 subgroup, with no overlap.
Comparison pathway analyses in IPA using eGenes [genes from cis-eQTL pairs unique to either CD1 or CD3 with p < 1e-08, FDR < 0.001] demonstrated that the CD3 subgroup was enriched in Wnt/beta-catenin signalling and regulation of epithelial–mesenchymal transition [Figure 5A], whereas the CD1 subgroup was enriched in pathways related to inflammation such as antigen presentation and OX40 signalling.
Figure 5.
cis-eQTL–based pathway analysis and transcriptional risk scores [TRSs] underline transition to increasing disease risk from CD1 to CD3. [A] Comparison pathway analysis using eGenes from cis-eQTLs unique to either CD1 or CD3 [p < 1e-08 for eQTL and p < 0.05 for Fisher’s exact test–based pathway analysis] revealed that the CD3 subgroup is enriched in Wnt/beta-catenin signalling and regulation of EMT. This possibly indicates that the underlying pathobiology for recurring and complicated disease in CD3 could be changes in epithelial architecture. [B] Association of TRSs with the three subgroups (p < 0.0001, Kruskal–Wallis [K–W] test). The CD3 subgroup was associated with higher TRSs compared with the CD1 subgroup, confirming CD3 as a more complicated and severely affected subset [p = 0.0002, Mann–Whitney [M–W] test].
3.6. Transcriptional risk scores
We calculated TRSs for the SB139 cohort using the methods described in the work by Marigorta et al.9 Transcriptional risk scores calculated using the expression data of the eGenes [eQTL-associated genes] in our SB139 cohort [see Methods] were found to be associated with the three CD subgroups [p < 0.0001, Kruskal–Wallis test] [Figure 5B]. The CD3 subgroup was associated with a significantly higher score compared with the CD1 subgroup [p = 0.0002, Mann–Whitney test]. The CD2 subgroup was intermediate, with a heterogeneous mix of subjects with both high and low TRS scores [Figure 5B]. Consistent with the conclusions from the Marigorta et al. study, the calculated TRSs in our study were positively associated with the subgroups with increasing disease severity.
3.7. Cell-type specific signatures were associated with subgroups
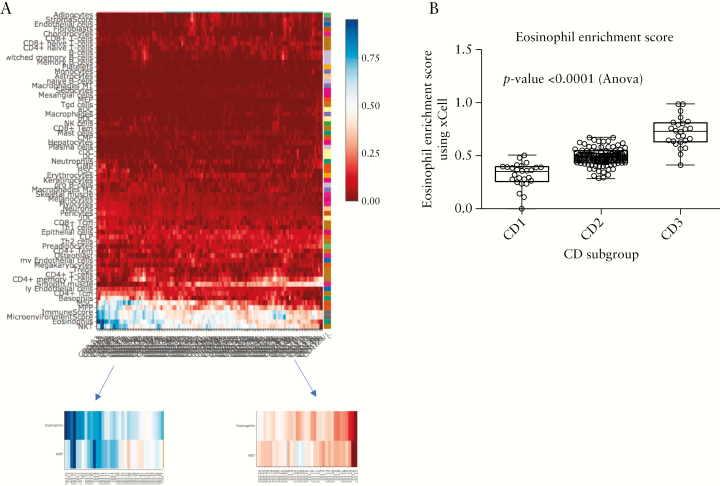
We examined the enrichment of the specific cell-type associated gene signatures in the SB resection tissue samples that could possibly indicate the subgroups we identified using the ileal tissue expression. We used xCell23 to generate cell-type-specific signatures associated with the three subgroups [Figure 6A]. The most pronounced cell-type differences, represented by the gene signature, were in the eosinophil and NKT enrichment scores, as highlighted in the left and right figure insets in Figure 6A. CD3 had a significantly higher eosinophil [EOS] enrichment score compared with the other subgroups [p < 0.0001] [Figure 6B]. We found that the NKT cell type enrichment scores were similarly associated with the subgroups [supplementary Figure S5]. To validate the presence of EOSs in the SB resected tissue, EOSs were manually counted on 67 of 139 H&E-stained sample slides [supplementary Figure S6]. We report the average EOS count per sample in supplementary Figure S6. We found that all the samples had EOSs present. However, we did not observe any statistically significant difference in the EOS counts across the three subgroups.
Figure 6.
Cell-type-specific signatures associated with the three subgroups. [A] Heat map showing the enrichment of different cell types in expression data from the uninvolved ileal tissue samples in the SB139 cohort obtained using the xCell23 tool. The left and right figure insets zoom in on the differential enrichment of eosinophil and NKT cells within the different samples. Higher enrichment scores were noticed in most of the samples belonging to the CD3 subgroup. [B] Statistical analysis of the eosinophil enrichment scores from xCell [ANOVA, p < 0.0001] indicated significantly higher scores in the CD3 subgroup and that CD1 had the lowest score. A similar trend was observed in NKT cells [supplementary Figure S5].
4. Discussion
Development of effective, personalized therapies for treating IBD and specifically CD patients has been hampered largely due to heterogeneity of clinical phenotypes, inaccurate patient stratification, and a lack of knowledge of associated pathways underlying the pathogenicity of each patient subclinical phenotype. We have attempted to address this issue by using a combination of genetics, transcriptomics, and clinical meta-data to interrogate the underlying pathogenesis of a severely affected CD population who had undergone a SB resection.
In this study, we analysed transcriptomic data from uninvolved ileal tissue samples from SB resections of CD patients [n = 139] and identified three patient subgroups, using multiple clustering algorithms. Subclinical phenotype associations indicated that the two most distant subgroups [CD1 and CD3] were clinically distinct and presented different disease courses. We focused on the extremes [CD1 and CD3 subgroups, 37% of the cohort] because we could associate these subgroups with distinct clinical phenotypes. This is consistent with previously reported work26 on medically refractory ulcerative colitis, in which only 20% of the cohort comprised the extreme subgroups associated with definitive risk of either having or not having colectomy.
Patients in the CD3 group were more severely affected compared with patients in CD1 and were associated with greater occurrence of second surgeries and shorter time to disease recurrence. The CD3 subgroup was also associated with pCD, a more severe form of CD. The mean TRS associated with the CD3 subgroup was found to be higher than those of CD1 and CD2. Our analysis identified CD3 as a more severely affected, homogenous subgroup among a population of heterogeneous CD patients who had undergone SB resection.
Gender differences have been reported to affect the prevalence of autoimmune diseases.27,28 Females have been found to be more predisposed to systemic diseases such as systemic lupus erythematosus, but prevalence is higher in males for rheumatological diseases such as ankylosing spondylitis. Women may be at higher risk of inflammation-associated diseases due to enhanced immune activation in the gut.29 Given these gender differences, enrichment of females in the CD3 group may indicate exacerbation of severe CD in females. However, the role of gender has not been carefully examined in IBD.
The CD2 subgroup was found to be heterogeneous and intermediate between CD1 and CD3, based on the TRS and survival analyses. This CD subtype did not have a clear subclinical phenotype and had a mix of pCD-positive and -negative patients. The time to recurrence from first surgery was found to be intermediate for this group. The TRS for the CD2 subgroup indicated a mix of low-risk [TRS < –1] as well as high-risk [TRS > 1] patients. However, we could not link any specific clinical phenotype with the CD2 patients based on TRS cut-offs of <–1 or >1. We speculate that the CD2 subgroup may represent a spectrum of phenotypes and, prospectively, these patients may be expected to transition to either of CD1 or CD3 characteristics. Our long-term goal is to eventually be able to stratify all patients, to identify the dominant pathway underlying their disease pathology and support the development of precision medicine.
Differential gene expression analyses comparing the CD1 and CD3 subgroups generated a list of gene expression signatures underlying the severe form of CD in the CD3 subgroup. Pathway analyses using this gene list revealed interesting sets of pathways that defined the poor disease course and the disease severity associated with CD3. The top pathway to be downregulated in the CD3 subgroup compared with CD1, with a negative activation z-score, was eIF2 signalling, among others such as Rho GTPases and actin signalling. Downregulated eIF2 signalling in the CD3 subgroup may be responsible for poor disease prognosis, because eIF2-related pathways are activated for robust autophagy responses to infection with CD-associated adherent–invasive Escherichia coli.30 Furthermore, mammalian cells with defective eIF2 signalling have been found to be more susceptible to bacterial invasion.31 The Rho family GTPases are known to be negatively regulated by Rho-specific guanine nucleotide dissociation inhibitor [RhoGDI] signalling.32 Consistent with this, we found RhoGDI signalling to be activated in the CD3 subgroup, whereas Rho GTPases signalling was downregulated in CD3. Rho GTPases connect external cellular signals to internal actin organization and in turn play a significant role in organization of actin cytoskeleton.33 We also found that the pathways related to actin signalling and regulation [actin-cytoskeleton signalling, regulation of actin-based motility by Rho] were downregulated in CD3 compared with CD1. We know that cell migration, and in turn wound healing, can be impaired by improper actin-cytoskeleton signalling; thus, we hypothesize that the CD3 subgroup of patients would have impaired wound healing compared with the CD1 patients.
Using a refined gene list for pathway analysis, consisting of genetic loci uniquely associated with either the CD1 or CD3 subgroups and also DE genes between the two subgroups, we revealed activation of the cAMP pathway in CD3. cAMP, the first intracellular second messenger, is known to play an important role in various signalling pathways associated with the pathogenesis of inflammatory diseases and is a potential therapeutic intervention point.34 Adenylate cyclase [AC] expression results in increased cAMP levels, whereas phosphodiesterase [PDE] activity inactivates cAMP. Protein kinase A [PKA] is activated by cAMP. Actin-based cell migration in turn is also regulated by the ‘cAMP/PKA’ signalling axis.35 cAMP/PKA signalling activity has been reported to affect actin cytoskeleton and cell migration both positively and negatively, and a balanced activity is believed to be important for successful cell migration. We identified activation of PKA signalling along with activated cAMP signalling in the CD3 subgroup. Consistent with a role for the cAMP/PKA pathway, phosphodiesterase 4C [PDE4C] was found to be overexpressed in the CD3 subgroup. Multiple SNPs at the PDE4C locus were found to be associated with the CD3 subgroup. This implies that PDE inhibitors present a logical choice among available therapeutics for patients in the CD3 subgroup. PDE inhibition is a known strategy for the treatment of autoimmune diseases, including IBD.36–38 Other differentially expressed genes that may be indicative of more severe disease, including intercellular adhesion molecule 3 [ICAM3], mothers against decapentaplegic homolog 3 [SMAD3], SH3, multiple ankyrin repeat domains 3 [SHANK3] and oncostatin-M receptor [OSMR]. ICAM-3 was found to be overexpressed in the CD3 subgroup and has been known to be associated with increased risk of having IBD in case control GWAS associations.39,40SMAD3 was downregulated in the CD3 subgroup, knock-out mice for SMAD3 have impaired intestinal mucosal healing,41 and SMAD3 variants have been shown to be associated with risk for recurring surgery in CD patients.42SHANK3 was downregulated in CD3, which is consistent with a reported role of SHANK3 in regulation of the intestinal barrier function, with SHANK3 knock-out mice showing an impaired epithelial barrier.43OSMR was upregulated in CD3, and high expression of oncostatin-M [OSM] and its receptor, OSMR, has been reported to be associated with disease severity and non-response to anti-TNF therapy.44 These findings provide a number of potential mechanisms by which the CD3 subgroup present with more severe disease, and generally indicate a heavily dysregulated gene signature that could predict disease severity.
Analysis of cell-type-specific signatures for our data predicted enhanced enrichment of EOS and NKT cell-type in CD3 compared with CD1, providing another possible mechanism for increased disease severity in the CD3 subgroup. However, manual EOS counts from H&E-stained tissue sections indicated that the enhanced EOS enrichment score predicted via xCell may be indicative of other cellular mechanisms such as EOS activation, rather than differential cell count, which needs to be further investigated. There has been speculation about the role of EOSs in IBD pathogenesis, because they are present in the gastrointestinal milieu and their possible interaction with other cells can impact epithelial barrier function and intestinal remodelling.45 Furthermore, a recent publication demonstrated that peripheral blood eosinophilia represented a biomarker of IBD patients at risk for poor clinical outcome.46
Our study has several limitations, including the relatively small sample size and unavailability of an appropriate independent validation cohort. In order to validate the genetic and transcriptomic signature associated with the refractory, severe CD3 subset, we would need an independent validation cohort of refractory CD patients. Another limitation is in the current possible use of this signature to identify patients that are at risk for complicated disease. As a future goal, we must seek to validate our findings and replicate them in peripheral blood in order for us to have a useable biomarker panel that can translate our findings from bench to clinic. In future, we also aim to conduct studies to increase understanding of tissue EOS activation status.
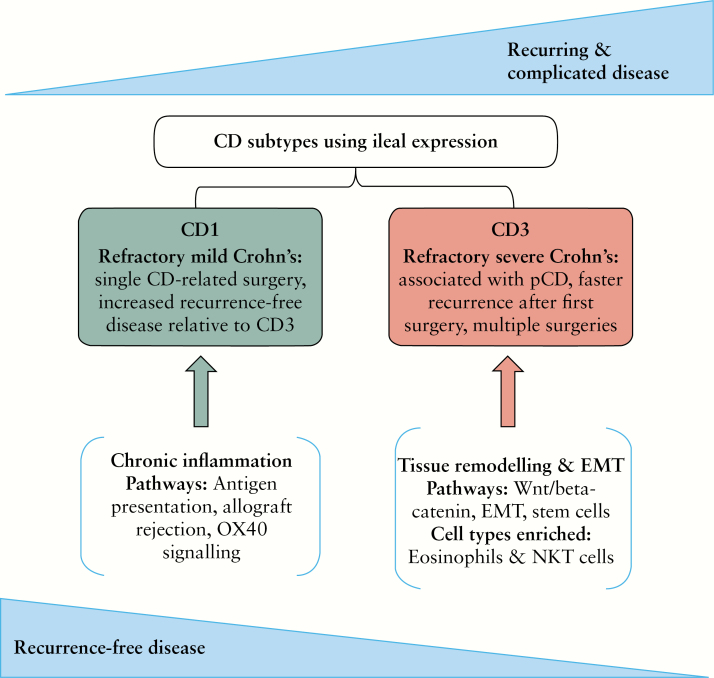
In conclusion, among a population of refractory CD patients that underwent SB resection, we identified CD3 as a more severely affected, refractory, distinct clinical subgroup [Figure 7]. Our genetic and transcriptomic analyses identified genetic burden, gene expression signature, and mechanistic pathways that could potentially underlie the pathogenesis of this severely affected patient subgroup. We have identified pathways and genetic signatures that might reflect an abnormality in wound healing, as indicated by the differential regulation of signatures for EMT and the WNT/B-catenin pathway. We’ve also identified signatures for infiltrating cell types that are implicated in intestinal remodelling. Finally, we have identified potential pathways that may provide clues as to the most appropriate therapeutic options for the patients that are faced with a poor quality of life and recurring, severe disease. Overall, in this study, we have demonstrated the advantage of using a multi-omic approach to the analysis of human disease to better inform the underlying pathobiology.
Figure 7.
Ileal expression from CD patients who underwent a single small bowel resection shows CD subtypes associated with varying extents of recurrence-free and complicated disease. We hypothesize that the CD1 and CD3 subgroups are located on the opposite ends of the refractory disease spectrum. Pathway analyses indicate the more severe CD3 subtype was involved in epithelial architecture changes driven by EMT [epithelial–mesenchymal transition], whereas CD1 was associated with chronic inflammation-related pathways.
Funding
This work was supported by internal funds from the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute.
Conflict of Interest
TH, KLV, TCL, TSS, PF: None; AAP: consults for Precision IBD; DL: consults for Precision IBD; SRT: consults for Precision IBD and is on Board of Directors for Robarts Clinical Trials; DPM: has consulted for Janssen, Pfizer, Gilead, Qu Biologics, Cidara, Precision IBD, and Bridge Therapeutics; MFF: consults for Precision IBD; and JB: consults for Precision IBD.
Supplementary Material
Acknowledgments
We are thankful to Carol Landers, Gregory Botwin, and the MIRIAD Biobank. The Cedars-Sinai MIRIAD IBD Biobank is supported by the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases [NIH/NIDDK] [grants P01 DK046763 and U01 DK062413], and The Leona M and Harry B Helmsley Charitable Trust.
Author’s Contributions
AAP: Study concept and design, literature search, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis, and technical support. DL: Acquisition of data, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, and statistical analysis. TH: Acquisition of data, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, and statistical analysis. KLV: Acquisition of data and revision of the manuscript. MFF: Acquisition of data. TCL: Acquisition of data and revision of the manuscript. PF: Acquisition of data. TSS: Acquisition of data. DPM: Study concept and design, acquisition of data, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, obtaining funding, and study supervision. SRT: Study concept and design, critical revision of the manuscript for important intellectual content, obtaining funding, and study supervision. JB: Study concept and design, literature search, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, obtaining funding, technical, or material support, and study supervision. All authors have read and approved this version as submitted.
References
- 1. Bilsborough J, Targan SR, Snapper SB. Therapeutic targets in inflammatory bowel disease: current and future. Am J Gastroenterol 2016;3:27–37. [Google Scholar]
- 2. Yoon SM, Haritunians T, Chhina S, et al. . Colonic phenotypes are associated with poorer response to anti-TNF therapies in patients with IBD. Inflamm Bowel Dis 2017;23:1382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell 2011;147:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bilsborough J, Dermot McGovern MP, Targan SR. Divide and conquer: using patient stratification to optimize therapeutic drug development in inflammatory bowel disease. J Immunol Clin Res 2014;2:1–4. [Google Scholar]
- 5. Gerich ME, McGovern DP. Towards personalized care in IBD. Nat Rev Gastroenterol Hepatol 2014;11:287–99. [DOI] [PubMed] [Google Scholar]
- 6. McGovern D. Personalized medicine in inflammatory bowel disease. Gastroenterol Hepatol 2014;10:662–4. [PMC free article] [PubMed] [Google Scholar]
- 7. Lee JC, Lyons PA, McKinney EF, et al. . Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 2011;121:4170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weiser M, Simon JM, Kochar B, et al. . Molecular classification of Crohn’s disease reveals two clinically relevant subtypes. Gut 2018;67:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marigorta UM, Denson LA, Hyams JS, et al. . Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat Genet 2017;49:1517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee JC, Biasci D, Roberts R, et al. ; UK IBD Genetics Consortium Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet 2017;49:262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. VanDussen KL, Liu TC, Li D, et al. . Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 2014;146:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. VanDussen KL, Stojmirović A, Li K, et al. . Abnormal small intestinal epithelial microvilli in patients with Crohn’s disease. Gastroenterology 2018;155:815–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ritchie ME, Phipson B, Wu D, et al. . Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009;19:1655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scrucca L, Fop M, Murphy TB, et al. . mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J 2016;8:147–65. [PMC free article] [PubMed] [Google Scholar]
- 16. Cleynen I, Boucher G, Jostins L, et al. ; International Inflammatory Bowel Disease Genetics Consortium Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC] Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–9. [DOI] [PubMed] [Google Scholar]
- 20. Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012;28:1353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC] Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. R Core Team. R: a language and environment for statistical computing [program]. Vienna, Austria: R Foundation for Statistical Computing; 2015. https://www.R-project.org/ Accessed August, 2018. [Google Scholar]
- 25. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haritunians T, Taylor KD, Targan SR, et al. . Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis 2010;16:1830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fairweather D, Frisancho-Kiss S, Rose NR. Sex differences in autoimmune disease from a pathological perspective. Am J Pathol 2008;173:600–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ngo ST, Steyn FJ, McCombe PA. Gender differences in autoimmune disease. Front Neuroendocrinol 2014;35:347–69. [DOI] [PubMed] [Google Scholar]
- 29. Sankaran-Walters S, Macal M, Grishina I, et al. . Sex differences matter in the gut: effect on mucosal immune activation and inflammation. Biol Sex Differ 2013;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nguyen H, Carriere J, Dalmasso G, et al. . OP002 The GCN2/eIF2α/ATF4 signaling pathway is necessary for autophagy response to infection with Crohn’s disease–associated adherent–invasive Escherichia coli. J Crohn’s Colitis 2014;8:S1–2. [Google Scholar]
- 31. Shrestha N, Bahnan W, Wiley DJ, Barber G, Fields KA, Schesser K. Eukaryotic initiation factor 2 [eIF2] signaling regulates proinflammatory cytokine expression and bacterial invasion. J Biol Chem 2012;287:28738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol 2011;12:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci 2011;124:679–83. [DOI] [PubMed] [Google Scholar]
- 34. Raker VK, Becker C, Steinbrink K. The cAMP pathway as therapeutic target in autoimmune and inflammatory diseases. Front Immunol 2016;7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Howe AK. Regulation of actin-based cell migration by cAMP/PKA. Biochim Biophys Acta 2004;1692:159–74. [DOI] [PubMed] [Google Scholar]
- 36. Spadaccini M, D’Alessio S, Peyrin-Biroulet L, et al. . PDE4 inhibition and inflammatory bowel disease: a novel therapeutic avenue. Int J Mol Sci 2017;18:1276–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kumar N, Goldminz AM, Kim N, Gottlieb AB. Phosphodiesterase 4–targeted treatments for autoimmune diseases. BMC Med 2013;11:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salari P, Abdollahi M. Phosphodiesterase inhibitors in inflammatory bowel disease. Expert Opin Investig Drugs 2012;21:261–4. [DOI] [PubMed] [Google Scholar]
- 39. Gu P, Theiss A, Han J, Feagins LA. Increased cell adhesion molecules, PECAM-1, ICAM-3, or VCAM-1, predict increased risk for flare in patients with quiescent inflammatory bowel disease. J Clin Gastroenterol 2017;51:522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vainer B, Nielsen OH. Changed colonic profile of P-selectin, platelet–endothelial cell adhesion molecule-1 [PECAM-1], intercellular adhesion molecule-1 [ICAM-1], ICAM-2, and ICAM-3 in inflammatory bowel disease. Clin Exp Immunol 2000;121:242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Owen CR, Yuan L, Basson MD. Smad3 knockout mice exhibit impaired intestinal mucosal healing. Lab Invest 2008;88:1101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fowler SA, Ananthakrishnan AN, Gardet A, et al. . SMAD3 gene variant is a risk factor for recurrent surgery in patients with Crohn’s disease. J Crohns Colitis 2014;8:845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei S-C, Yang-Yen H-F, Tsao P-N, et al. . SHANK3 regulates intestinal barrier function through modulating ZO-1 expression through the PKCε-dependent pathway. Inflamm Bowel Dis 2017;1–11. [DOI] [PubMed] [Google Scholar]
- 44. West NR, Hegazy AN, Owens BMJ, et al. ; Oxford IBD Cohort Investigators Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woodruff SA, Masterson JC, Fillon S, Robinson ZD, Furuta GT. Role of eosinophils in inflammatory bowel and gastrointestinal diseases. J Pediatr Gastroenterol Nutr 2011;52:650–61. [DOI] [PubMed] [Google Scholar]
- 46. Click B, Anderson AM, Koutroubakis IE, et al. . Peripheral eosinophilia in patients with inflammatory bowel disease defines an aggressive disease phenotype. Am J Gastroenterol 2017;112:1849–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.