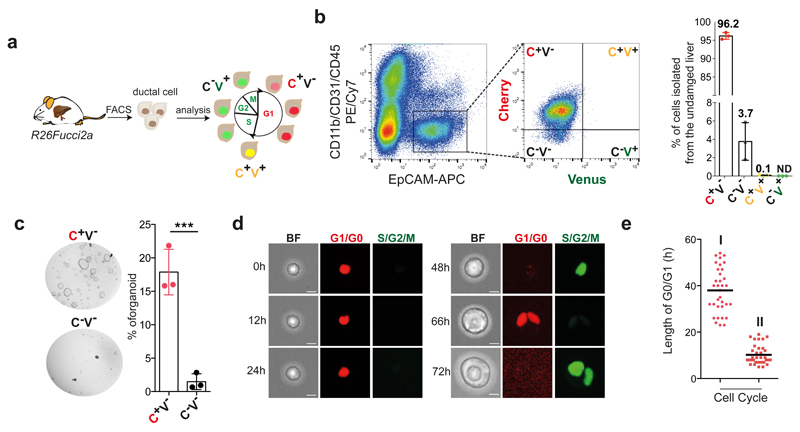
Fig. 1. G1/G0 arrested liver ductal cells require ~48h to start cell proliferation and initiate liver organoids cultures.
R26Fucci2a mice constitutively express a bi-cistronic cell-cycle reporter that allows discriminating between G1/G0 [Cherry-hCdt1+ (30/120), red] and S/G2/M [Venus-hGem+, (1/110) green] phases of the cell cycle. a, Experimental approach b, EpCAM+ liver ductal cells from R26Fucci2a mice were FACS-sorted according to the expression of mCherry-hCdt1 (C) and/or mVenus-hGem (V). The graph represents percentage of EpCAM+ cells positive for mCherry and/or mVenus. Each dot represents an independent experiment from an independent mouse (n=3). Graph is presented as mean±SD of 3 independent experiments. c, Representative bright field images of 500 C+/V- EpCAM+ and C-/V- EpCAM+ cells cultured for 6 days as liver organoids. The graph represents mean±SD of organoid formation efficiency (n=3 experiments). **, p-value (p=0.001413095) was calculated using Student's two tailed t-test. **, p<0.01. d, Still images from a representative movie of C+/V- EpCAM+ ductal cells monitored for 72h using a spinning-disk confocal microscope. Scale bars, 10μm. e, Graph represents G0/G1 length for the first (I) and second (II) cell cycles since t=0h (isolation) of 34 cells (n=3 independent experiments). Global mean of G0/G1 length is shown (G0O/G1 I = 37.97h hours; G0O/G1 II = 10.20h hours). h, hours.