Abstract
The antagonistic anti-CD40 antibody, 2C10, and its recombinant primate derivative, 2C10R4, are potent immunosuppressive antibodies whose utility in allo- and xenotransplantation have been demonstrated in nonhuman primate studies. In this study, we defined the 2C10 binding epitope and found only slight differences in affinity of 2C10 for CD40 derived from four primate species. Staining of truncation mutants mapped the 2C10 binding epitope to the N-terminal portion of CD40. Alanine scanning mutagenesis of the first 60 residues in the CD40 ectodomain highlighted key amino acids important for binding of 2C10, and for binding of the noncross-blocking anti-CD40 antibodies 3A8 and 5D12. All four 2C10-binding residues defined by mutagenesis clustered near the membrane-distal tip of CD40 and partially overlap the CD154 binding surface. In contrast, the overlapping 3A8 and 5D12 epitopes map to an opposing surface away from the CD154 binding domain. This biochemical characterization of 2C10 confirms the validity of nonhuman primate studies in the translation of this therapeutic antibody and provides insight its mechanism of action.
1. Introduction
CD40 (Tumor Necrosis Factor Receptor Superfamily 5, TNFRS5) is a transmembrane protein expressed on diverse immune cell types, including B lymphocytes, monocytes, and macrophages. In addition to its constitutive expression on various immune cells, CD40 has also been detected on non-lymphoid tissues such as endothelial and smooth muscle cells during inflammatory states. Activation of CD40 on B cells by interaction with ligand CD154 expressed on T lymphocytes promotes activation, germinal center formation, class switching, and somatic maturation. Downstream effects of CD40-CD154 engagement are mediated primarily by TRAF proteins that directly or indirectly engage clustered CD40 cytoplasmic domains and lead to, among other activation events, signaling through NF-kB (1).
The CD40-CD154 interaction is a target of interest in transplantation to improve long-term graft survival, broaden donor-recipient pairings, and overcome xenotransplantation hurdles related to rejection. Furthermore, targeting of this pathway offers a promising approach with utility in calcineurin-free treatment regimens (2–5). A number of anti-CD40 antibodies have been tested in murine and primate models of transplantation (4, 6). We have previously described 2C10R4, a mouse-rhesus IgG4 chimeric antibody, as having purely antagonist, non-depleting, and ligand blocking anti-CD40 activity (7). Administration of 2C10R4 for induction and maintenance resulted in long-term graft survival of engineered porcine cardiac xenografts transplanted into baboons both heterotopically (8) and orthotopically (9). Favorable outcomes were also observed using 2C10R4 immunosuppression for kidney (10, 11) and cornea (12) xenografts in nonhuman primates attesting further to its translational potential.
Antibodies targeting CD40 are under development for several clinical applications, including as immunosuppressive therapeutics or inflammatory antitumor agents, and it is believed that the capacity to block or activate is highly influenced by the epitope location. In this study, we have defined the 2C10 binding epitope and compared it to the epitope of related, but noncross-blocking agonist anti-CD40 monoclonal antibodies (mAbs) 3A8 and 5D12. (13). All three mAbs bind to the membrane-distal tip of CD40 near the CD154-interacting surface, but only 2C10 competes for ligand binding. In this study, a mutagenesis analysis coupled with protein modeling provides insight into the antibodies’ mechanisms of action and illustrates the profound impact of even subtle epitope differences on target cell biology.
2. Materials and Methods
2.1. Generation of CD40 expression constructs
Rhesus macaque (M. mulatta) cDNA was generated from isolated PBMC RNA by Superscript III (Thermo Fisher) and an oligo (dT)15–18 primer (Roche) using standard conditions outlined by the manufacturer. Primers homologous to the predicted CD40 (AA 1–277) sequence (XM_015148819.1) were designed to amplify the full-length transcript using Phusion High Fidelity DNA polymerase (New England Biolabs) under standard conditions. Pooled amplicons were sequenced and subcloned into pcDNA3.1 (+) (Thermo Fisher). The final sequence was deposited into NCBI (MF498903) and includes two non-silent polymorphisms compared to the described M. mulatta genomic assembly.
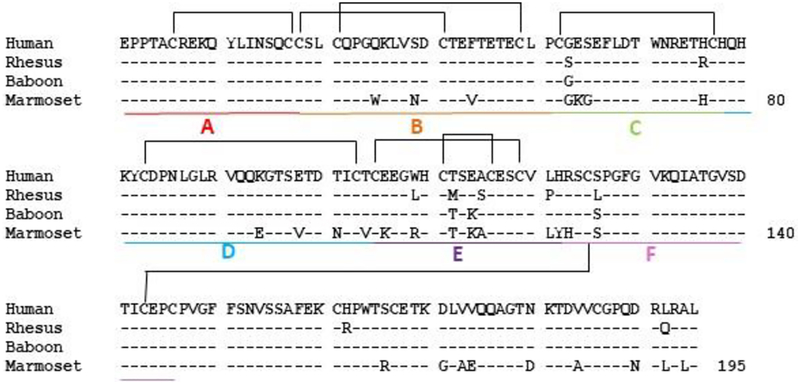
A His-tagged, soluble CD40 (sCD40-His) expression construct was generated encoding the extracellular domain of macaque CD40 (AA 21–193) and subcloned into the pcDNA™3.4 TOPO® expression vector (Thermo Fisher). Published sequences for human (H. sapiens, sequence ID: AAO43990.1), baboon (P. anubis, NP_001306202.1), and marmoset (C. jacchus, NP_001254659.1) CD40 were referenced to generate Figure 1 and analogous sCD40-His constructs were created for each primate species.
Figure 1:
Alignment of primate CD40. Amino acid sequences from human (acc. no. AA043990.1), rhesus macaque (acc. no. MF498903), olive baboon (acc. no. NP_001306202.1), and common marmoset (acc. no. NP_001254659.1) CD40 ectodomains are shown. Disulfide linkages are bracketed and domain modules analyzed in Figure S1 are labeled A-F.
Full length CD40 was cloned into pcDNA3.1(+) and used as a template for the generation of 48 unique alanine mutants. Mutagenesis reactions were carried out using the Q5® Site-Directed Mutagenesis Kit (New England Biolabs) according to the manufacturer’s instructions.
2.2. Expression and purification of soluble CD40-His proteins
Recombinant sCD40-His proteins were produced using the ExpiCHO™ Expression System (Thermo Fisher Scientific) according to the manufacturer’s protocol. At day 5 post transfection, supernatant containing soluble CD40 was clarified by centrifugation and filtration. Cell culture supernatants containing sCD40-His were applied to a 1 mL HisTrap™ and purified according to the manufacturer’s instructions. 20 mM and 250 mM imidazole were used in the wash and elution buffers, respectively. Eluates were concentrated and exchanged into PBS before being applied to a 10x300 mm Superdex 200 Increase gel filtration column (GE Healthcare) for further purification. Fractions containing recombinant sCD40-His were pooled and exchanged into HBS-EP+ buffer (0.01M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20 (GE Healthcare).
2.3. Determination of antibody kinetic rate constants by surface plasmon resonance
The mouse anti-CD40 antibodies 2C10 (15), 3A8 (ATCC) and 5D12 (ATCC) were expressed from hybridomas. The mouse-rhesus IgG4 chimeric anti-CD40 (2C10R4) and mouse-rhesus IgG1 chimeric (2C10R1), were expressed from stably transduced CHO cells. All were grown in serum-free medium and purified by protein A affinity chromatography.
Kinetic analysis of antibody-antigen interactions was performed on the Biacore T200 (GE Healthcare) using the ligand capture approach. For all Biacore experiments, HBS-EP+ buffer was used as the running and sample buffer unless otherwise noted.
For kinetics assays, 75 RU of 2C10R1 was captured onto flow cell 2 at a flow rate of 10 μL/minute. Following antibody capture, a dilution series of sCD40-His was injected over flow cells 1 and 2 at 50 μL/minute. Injections of 100, 50, 25, 12.5, 6.25 and 3.125 nM sCD40-His were applied, and association and dissociation was monitored for 300 and 600 seconds, respectively. The sensor surface was regenerated using 3M MgCl2. The 12.5 nM injection was repeated at the end of each kinetics series to monitor drift within individual experiments. For kinetic characterization of soluble CD40 alanine mutants, analyte was injected at the concentrations indicated in Figure 4 (2C10) and Figure S4 (5D12).
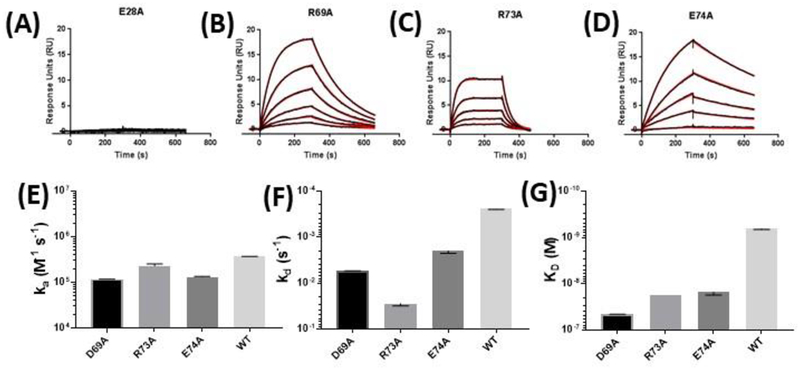
Figure 4:
Binding kinetics of select rhesus sCD40 alanine mutants. Purified sCD40 mutants E28A (A), D69A (B), R73A (C), and E74A (D) were flowed over immobilized 2C10 and the kinetics of the interaction were characterized by SPR. Analyte injections of 80, 40, 20, 10, 5.0 and 2.5 nM were tested for each mutant. Kinetic rate constants could not be determined for the 2C10-E28A interaction, as no detectible binding was observed at concentrations as high as 500 nM (data not shown). Measurable calculated ka (E), kd (F), and KD (G) are compared to wildtype for D69A, R73A, and E74A. All mutant and wildtype protein preparations were first checked for conformation integrity by CFCA (30) and affinity measurements are corrected for active protein concentrations. Kinetic value plots represent at least three independent measurements. Error bars represent SEM.
All kinetics experiments were performed in triplicate and reported rate constants reflect the average of 3 separate runs. SPR data were analyzed with Biacore T200 Evaluation software and kinetic rate constants were determined using the 1:1 Langmuir binding model describing 1:1 binding between the ligand (mAb) and analyte (sCD40-His).
2.4. α-CD40 domain mapping
24 hours prior to transfection, HEK293T cells (ATCC) were seeded in 6-well plates at 2.0 × 105 cells/well containing Gibco™ DMEM plus 10% FBS and cultured at 37°C with 5% CO2. FLAG-tagged CD40 constructs or pcDNA3.1 empty vector were transfected into cells using Lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s protocol. At 48 hours post-transfection, cells were stained with anti-FLAG-APC (Miltenyi Biotech) and ⍺-CD40 antibodies 2C10-PE (1.25 μg/test), 3A8-FITC (0.50 μg/test) or 5D12-Alexa488 (0.25 μg/test). All flow cytometry data was collected on the MACSQuant® Analyzer 10 (Miltenyi Biotech).
2.5. α-CD40 mAb competition assay
2C10R1 was captured onto the anti-human IgG surface of a CM5 chip to a response level of 300 RU. After 2C10R1 capture, a 60 second injection of 500 nM rhesus sCD40-His was performed over flow cells 1 and 2, with flow cell 1 serving as the reference cell. After analyte binding, 100 nM of mouse 2C10, 3A8, or 5D12 was applied to both flow cells and epitope competition was evaluated based on the presence or absence of a secondary association event. At the end of each cycle, the sensor surface was regenerated with a 30 second injection of 3M MgCl2. All steps were carried out at a flow rate of 10 μL/minute using HBS-EP+ as the running and sample buffer.
2.6. Transient transfection of CD40 alanine mutants
One day prior to transfection with the CD40 alanine mutant panel, 96-well tissue-culture microplates were seeded with 10,000 HEK293T cells per well (DMEM + 10% FBS). For each well to be transfected, 200 ng of plasmid DNA containing CD40 mutants or control was transfected using Lipofectamine 3000 (Thermo Fisher) following the manufacturer’s recommendations. Cells were cultured at 37°C for 48 hours before being analyzed by flow cytometry.
2.7. Epitope mapping assay
Immunoreactivity of 2C10, 3A8, and 5D12 against the CD40 alanine mutant panel was measured by flow cytometry. 2C10, 3A8, and 5D12 were conjugated to PE, FITC, and Alexa Fluor 488, respectively. Cells were stained with 12.5 μg/mL 2C10, 5.0 μg/mL 3A8, and 2.5 μg/mL 5D12 for 30 minutes before being washed with 2mM EDTA in dPBS. Finally, samples were fixed in 2% formaldehyde before being analyzed by flow cytometry using a MACSQuant® Analyzer 10 (Miltenyi Biotech).
2.8. Statistical analysis
Data are shown as the mean ± standard error of the mean calculated in Excel. Linear regression with 95% confidence intervals was determined using Graphpad Prism.
3. Results
3.1. Species comparison of 2C10-CD40 binding.
Anti-CD40–2C10 was originally generated by immunizing mice with rhesus soluble CD40 (sCD40) and screening resulting hybridomas against human sCD40 (7). We performed a comparative analysis between rhesus macaque, human, baboon, and marmoset sCD40 to confirm the translational relevance of Old and New World primate models to evaluate 2C10 treatment regimens. In a protein alignment, rhesus and human sCD40 share 95% sequence similarity, while baboon is 98% similar to rhesus (Figure 1). As expected, New World primate species marmoset was the most distantly related with only 83–87% sequence similarity to the other species.
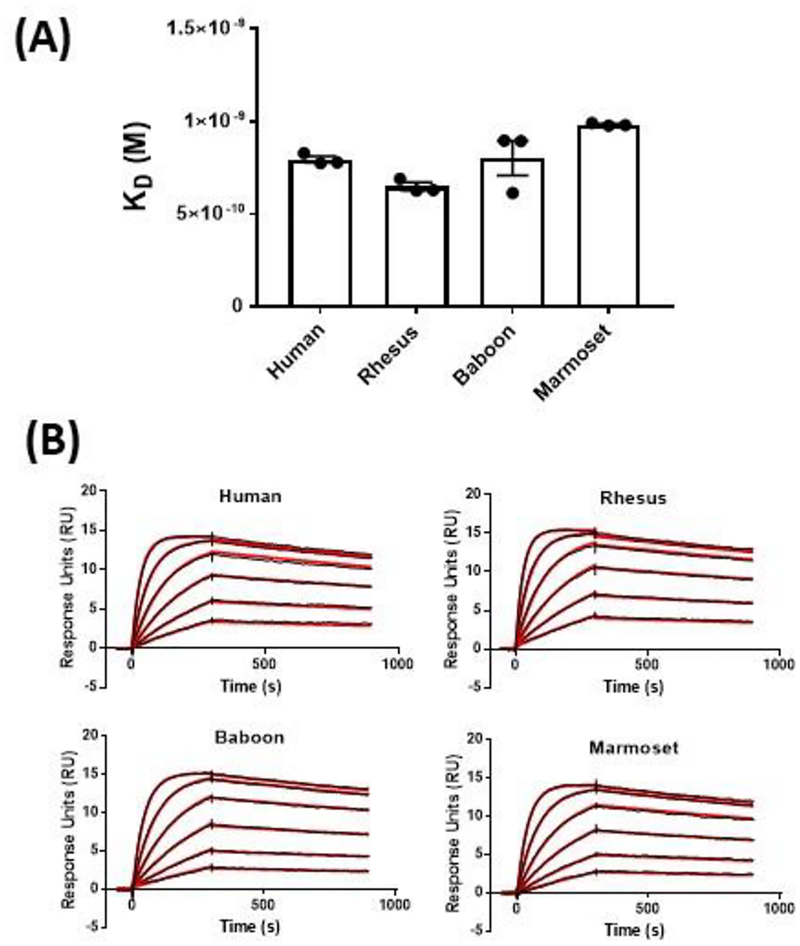
To compare the binding affinity of primate CD40 molecules to 2C10, SPR analysis was performed using purified recombinant protein from each species. Representative sensorgrams of the species comparison are shown in Figure 2B. As seen in Figure 2A, relative binding affinities between primate sCD40 were similar in magnitude with all KD measurements falling between 0.65 and 1.0 nM. As suggested in a previous report that compared staining levels in PBMC, affinity of 2C10 was slightly lower for baboon than rhesus CD40.
Figure 2:
Comparison of 2C10 binding to human, rhesus macaque, baboon, and marmoset CD40. Injections of 100, 50, 25, 12.5, 6.25 and 3.125 nM sCD40-His were applied to sensor surface containing 75 RU of 2C10R1 and association and dissociation was monitored for 300 and 600 seconds, respectively. Relative conformational integrity of all four sCD40s was verified by calibration-free concentration analysis (CFCA) (30). Representative sensorgrams with raw values in black and kinetic fit in red for each species are included in (A). The mean KD values from three independent measurements by SPR analysis of primate sCD40 and 2C10 are shown in (B). Error bars represent SEM from 3 experiments.
3.2. Refining the 2C10 epitope.
Like other TNFR family proteins, the extracellular domain of CD40 is composed of 175 amino acids arranged in a ladder-like structure with 4 cysteine-rich domains (CRDs) that each contain 2 or more disulfide bonds (Figure 1) (14). In order to narrow the potential 2C10-interaction region, several N-terminal truncation mutants, guided by the CRD boundaries to minimize profound conformation shifts, were generated and transfected into HEK293T cells for flow analysis (Figure S1 and Table S1). Each protein was tagged with an N-terminal FLAG epitope to correct for surface expression differences. 2C10 staining decreased below detectable levels after truncation of the first 17 N-terminal amino acids, indicating removal of one or more critical binding residues. Binding of 3A8 and 5D12 also significantly declined with truncation BCDEF (residues 38–195), but residual staining suggests that the epitope for these antibodies is less dependent on the first 17 amino acids than for 2C10.
3.3. Alanine scanning mutagenesis of the CD40 N-terminus.
In order to map exact 2C10 interacting residues, single alanine or glycine substitutions were introduced into the first 60 amino acids of the full length rhesus macaque protein (Figure 1). Cysteines and prolines were excluded to minimize disruptions to conformation. Each point mutant was transfected into 293T cells and stained with 2C10, 3A8, or 5D12 for analysis by flow cytometry. To first verify that 2C10 and 3A8 or 5D12 occupy non-overlapping epitopes, a competition assay was performed by SPR that detected dual binding of 2C10 and either 3A8 or 5D12 to sCD40, indicating that these antibodies do not cross-block (Figure S2).
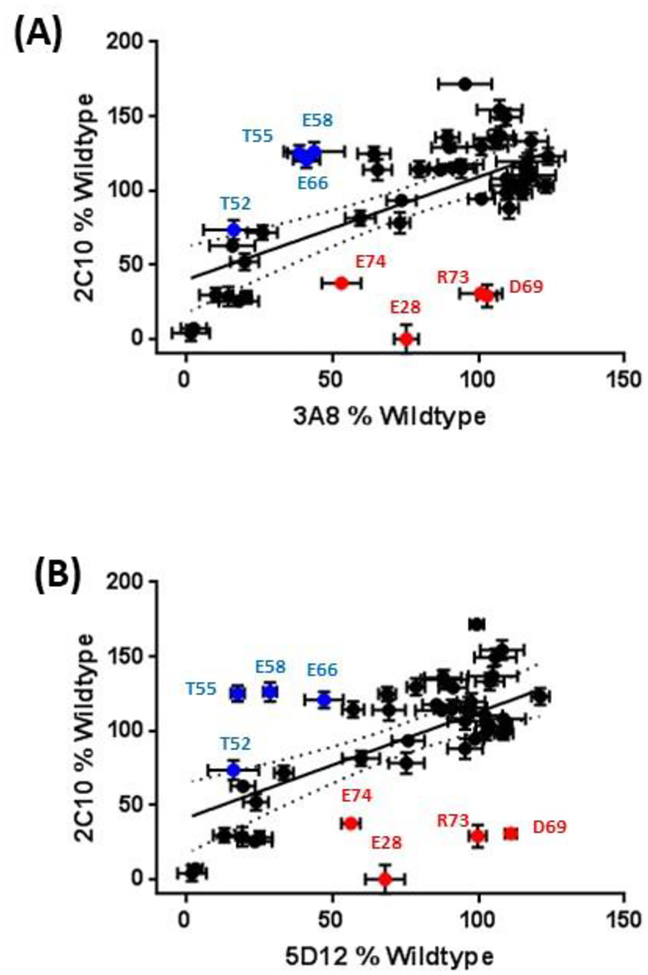
Significant shifts in fluorescence intensity occurred when positions E28, D69, R73, and E74 were mutated to alanine, and these changes were unique to 2C10 (Figure S3). The dot plots in Figure 3 further illustrates the selective reduction in 2C10 staining at these positions compared to 5D12 and 3A8. 5D12 and 3A8 staining was likewise selectively reduced in mutants T52A, T55A, E58A, and E66A, indicating that these residues partially comprise a shared epitope.
Figure 3:
Alanine scan summary. Staining of each alanine mutant by 2C10 is expressed as the percent geometric mean fluorescence intensity (gMFI) relative to wildtype and plotted against the analogous measurement with non-competing 3A8 (A) or 5D12 (B). Each experiment was repeated in triplicate with error bars representing the SEM. Red plot points represent mutants that fell below the 95% confidence interval of the regression slope and showed a 2C10:3A8 or 2C10:5D12 ratio of ≤1:1.5; blue plot points indicate mutants with 3A8:2C10 or 5D12:2C10 ratio of ≤1:1.5.
When expressed as soluble mutants, E28A, D69A, R73A, and E74A also demonstrated lower affinity for 2C10 by SPR analysis, with the most profound decrease in binding observed from E28A (Figure 4). E28A completely abrogated 2C10 but not 5D12 binding (Figure 4A and S4), confirming the results obtained from the flow cytometry assay (Figure 4). Notably, changes in 2C10 affinity for each mutant were mostly due to the increased dissociation rate (kd), shown in Figure 4F.
3.4. Epitope structure mapping.
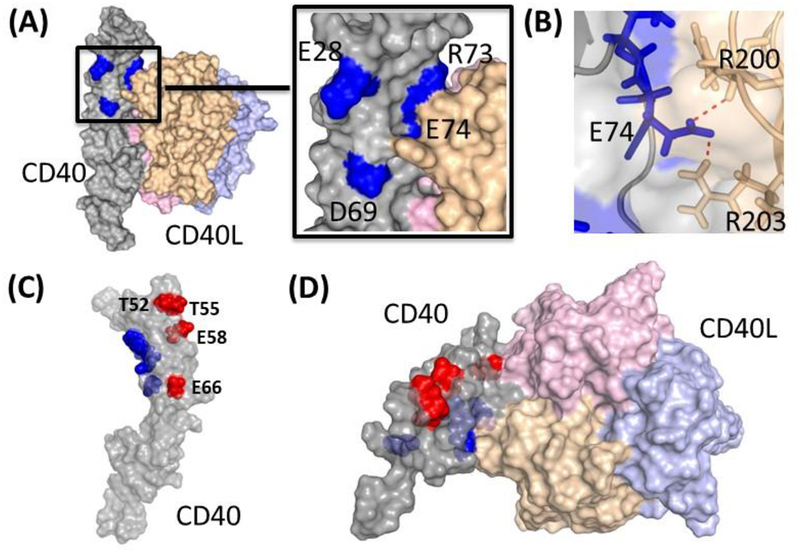
The CD40-CD154 interaction occurs in a 2:3 molar ratio with two CD40 molecules binding asymmetrically to the CD154 trimer. The N-terminal CRD1 and CRD2A make similar, mostly charged, contacts with CD154 on both surfaces, while the entire CRD1–3 span interacts on the other surface (15). To visualize our findings in PyMOL (Schrodinger), we used a previously resolved CD40-CD154 complex structure (PDB 3QD6) (Figure 5). All four critical residues identified in this study – E28, D69, R73, and E74 – cluster near the membrane-distal tip of CD40 (Figure 5A). Residues E28 and D69 in CRD1 sit orthogonal and adjacent to the CD154 binding interface, while R73 and E74 both directly interact with CD154. It seems that the high affinity interaction between 2C10 and E28 positions the antibody squarely in CRD1, but weaker interactions in or near the CD154 interface may angle it over the ligand-binding surface.
Figure 5:
Crystal structure of the human CD40-CD154 complex generated from published structure data (Protein Data Bank accession code 3QD6). (A) Surface representation of CD40 (gray, with the four critical residues -E28, D69, R73 and E74- required for 2C10 binding shown in blue) and the CD154 trimer (consisting of three subunits, in pink, yellow and violet); inset, magnification of the CD40/CD154 contact surface around the four critical residues. (B) Rotated magnification of the CD40/CD154 contact surface around the residue E74 of CD40. Charge interactions are depicted as red dashed lines between CD40 (residue E74 represented as blue sticks) and CD154 (residues R200 and R203 represented as yellow sticks). (C) CD40 structure showing in blue the relative positions of the 2C10 epitopes and in red the 5D12/3A8 epitopes (T52, T55, E58 and E66). (D) Top view of the CD40/CD154 structure showing the relative positions of the 2C10 (blue) and 5D12/3A8 (red) epitopes.
Results obtained from SPR competition and alanine-scanning mutagenesis experiments suggest that 3A8 and 5D12 have overlapping and nearly identical epitopes. Sequencing and alignment of heavy and light chain variable regions revealed only a single amino acid difference between 5D12 and 3A8 CDRs, indicating that they are likely derived from the same lineage (data not shown). In contrast to 2C10, the 3A8 and 5D12 epitopes span from the membrane-distal tip to midway down the molecule’s edge on an opposing face (Figure 5C and D).
4. Discussion
In this study, we describe the epitopes for the anti-CD40 antagonist antibody 2C10 as well as the mildly agonist antibodies 5D12 and 3A8. We also compared the CD40 sequence homology and relative 2C10 binding affinity between human, baboon, rhesus macaque and marmoset CD40. All four amino acids comprising the 2C10 epitope were conserved in the cercopithecine monkeys, and three of four were conserved in marmosets. As expected, the binding affinities of 2C10 for non-human primate CD40, when measured by SPR, did not differ significantly among all primate species tested. The binding affinities of 2C10 for non-human primate CD40, when measured by SPR differed by up to 20%. This conclusion deviates from that of Lee et. al. who showed dimmer staining of baboon B cells by 2C10 than macaque or human B cells by ~2-fold (18). However, we believe SPR provides a more accurate measurement of antibody binding affinity versus cell staining of PBMC where it is not possible to account for the number of CD40 molecules on the cell surface. Furthermore, unlike the cytofluorimetric approach cited above, this technique enables the comparison of interaction kinetics in real-time. The results of Lee et. al. do not rule out subtle difference in CD40 biology between primate species, such as level of CD40 expression on circulating B cells. Nevertheless, our finding of epitope conservation and similar binding affinity support the relevance of macaque and baboon studies in the translation of 2C10 as a drug with indications for blocking transplant rejection and other autoimmune diseases.
A number of other antagonistic anti-CD40 antibodies have shown success in allograft and xenograft survival in NHP or have advanced to clinical trials as candidate immunosuppressive agents. Chi220 was effective in maintaining kidney allografts in nonhuman primates (16). Antibodies targeting the 3A8/5D12 epitope suppressed allograft rejection in nonhuman primates and showed effect in modulating autoimmune gastrointestinal inflammation in a phase II clinical trial (17–19). Both ASKP1240 and CFZ533 have demonstrated safety and clinical effectiveness in autoimmune or transplantation indications (20–24). BI655064 is being evaluated clinically in patients with rheumatoid arthritis (25). Most of these anti-CD40 antibodies were selected for antagonistic activity and an absence of cell-depleting characteristics. However, their precise mechanism of action remains unclear.
Mapping of the 2C10 epitope to the co-crystal structure of CD40 and CD154, shows that the binding residues cluster near the membrane-distal tip of CD40 which has the potential to interfere with CD40 interacting with its ligand, CD154. Indeed, early characterization of 2C10 activity in vivo showed its ability to completely block binding of CD154 to B cells (15). Likewise, partially agonistic anti-CD40 antibody Chi220’s epitope overlaps the CD154 binding surface, blocks engagement, but does not compete with 2C10 for binding (17, 26). One study linked the position of antibody binding to the CD154-binding surface to agonist activity (27). However, antibodies targeting the 3A8/5D12 epitope, while clearly immunosuppressive, do not directly interfere with CD40 interacting with its ligand. These antibodies appear to alter TRAF signaling at a downstream location (28).
Another study linked epitope position relative to membrane proximity as predictive of CD40 antibody agonist, antagonist, and CD154-blocking activity (29). In this analysis, antibodies that bind CRD 2–4 were more likely than membrane-distal CRD1 to function both as antagonist and CD154-blocking. However, the data presented here suggests that CD40 antibody function is more nuanced and CRD1-binding antibodies can also function antagonistically. Further studies elucidating antibody mechanisms of action may clarify factors that firmly contribute to function of antibodies targeting CD40 and other TRAF-signaling co-stimulatory proteins.
Supplementary Material
Acknowledgments
This work was supported by grant number U24AI26683 from the National Institute of Allergy and Infectious Diseases.
Abbreviations
- TNFRSF
Tumor Necrosis Factor Receptor Super Family
- TRAF
Tumor Necrosis Factor Receptor Associated Protein
- NF
Nuclear Factor
- PBMC
Peripheral Blood Mononuclear Cells
- cDNA
Complementary Deoxyribose Nucleic Acid
- HBS-EP+
0.01M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20
- ATCC
American Tissue Culture Company
- CHO
Chinese Hamster Ovary
- HEK
Human Embryotic Kidney
- RU
Response Units
- SPR
Surface Plasmon Resonance
- sCD40
Soluble (Extracellular domain) CD40
- CM5
Carboxymethylated Dextran
- EDTA
Ethylenediaminetetraacetic Acid
- dPBS
Dulbecco’s Phosphate-buffered Saline
- CFCA
Calibration-free Concentration Analysis
- CRD
Cysteine-rich Domain
- CDR
Complementary-determining Region
- KLH
Keyhole Limpet Hemocyanin
Footnotes
Disclosure
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. K.A.R owns stock in Kiniksa Pharmaceuticals which has licensed certain rights to the 2C10 antibodies described herein. The other authors have no conflicts of interest to disclose.
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- 1.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 2009;229(1):152–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill L, Alousi A, Kebriaei P, Mehta R, Rezvani K, Shpall E. New and emerging therapies for acute and chronic graft versus host disease. Ther Adv Hematol 2018;9(1):21–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matas AJ. Minimization of steroids in kidney transplantation. Transpl Int 2009;22(1):38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinelli DF, Ford ML. Novel insights into anti-CD40/CD154 immunotherapy in transplant tolerance. Immunotherapy 2015;7(4):399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porrini E, Moreno JM, Osuna A, Benitez R, Lampreabe I, Diaz JM et al. Prediabetes in patients receiving tacrolimus in the first year after kidney transplantation: a prospective and multicenter study. Transplantation 2008;85(8):1133–1138. [DOI] [PubMed] [Google Scholar]
- 6.Zhang T, Pierson RN 3rd, Azimzadeh AM. Update on CD40 and CD154 blockade in transplant models. Immunotherapy 2015;7(8):899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe M, Badell IR, Thompson P, Martin B, Leopardi F, Strobert E et al. A novel monoclonal antibody to CD40 prolongs islet allograft survival. Am J Transplant 2012;12(8):2079–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohiuddin MM, Singh AK, Corcoran PC, Thomas ML 3rd, Clark T, Lewis BG et al. Chimeric 2C10R4 anti-CD40 antibody therapy is critical for long-term survival of GTKO.hCD46.hTBM pig-to-primate cardiac xenograft. Nat Commun 2016;7:11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langin M, Mayr T, Reichart B, Michel S, Buchholz S, Guethoff S et al. Consistent success in life-supporting porcine cardiac xenotransplantation. Nature 2018;564(7736):430–433. [DOI] [PubMed] [Google Scholar]
- 10.Iwase H, Ekser B, Satyananda V, Zhou H, Hara H, Bajona P et al. Initial in vivo experience of pig artery patch transplantation in baboons using mutant MHC (CIITA-DN) pigs. Transpl Immunol 2015;32(2):99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwase H, Liu H, Wijkstrom M, Zhou H, Singh J, Hara H et al. Pig kidney graft survival in a baboon for 136 days: longest life-supporting organ graft survival to date. Xenotransplantation 2015;22(4):302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Kim DH, Choi HJ, Lee HJ, Kang HJ, Park CG et al. Anti-CD40 antibody-mediated costimulation blockade promotes long-term survival of deep-lamellar porcine corneal grafts in non-human primates. Xenotransplantation 2017;24(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Boer M, Conroy L, Min HY, Kwekkeboom J. Generation of monoclonal antibodies to human lymphocyte cell surface antigens using insect cells expressing recombinant proteins. J Immunol Methods 1992;152(1):15–23. [DOI] [PubMed] [Google Scholar]
- 14.Naismith JH, Sprang SR. Modularity in the TNF-receptor family. Trends Biochem Sci 1998;23(2):74–79. [DOI] [PubMed] [Google Scholar]
- 15.An HJ, Kim YJ, Song DH, Park BS, Kim HM, Lee JD et al. Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J Biol Chem 2011;286(13):11226–11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson TC, Trambley J, Odom K, Anderson DC, Cowan S, Bray R et al. Anti-CD40 therapy extends renal allograft survival in rhesus macaques. Transplantation 2002;74(7):933–940. [DOI] [PubMed] [Google Scholar]
- 17.Badell IR, Thompson PW, Turner AP, Russell MC, Avila JG, Cano JA et al. Nondepleting anti-CD40-based therapy prolongs allograft survival in nonhuman primates. Am J Transplant 2012;12(1):126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boon L, Laman JD, Ortiz-Buijsse A, den Hartog MT, Hoffenberg S, Liu P et al. Preclinical assessment of anti-CD40 Mab 5D12 in cynomolgus monkeys. Toxicology 2002;174(1):53–65. [DOI] [PubMed] [Google Scholar]
- 19.Kasran A, Boon L, Wortel CH, Hogezand RA, Schreiber S, Goldin E et al. Safety and tolerability of antagonist anti-human CD40 Mab ch5D12 in patients with moderate to severe Crohn’s disease. Aliment Pharmacol Ther 2005;22(2):111–122. [DOI] [PubMed] [Google Scholar]
- 20.Anil Kumar MS, Papp K, Tainaka R, Valluri U, Wang X, Zhu T et al. Randomized, controlled study of bleselumab (ASKP1240) pharmacokinetics and safety in patients with moderate-to-severe plaque psoriasis. Biopharm Drug Dispos 2018;39(5):245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldwater R, Keirns J, Blahunka P, First R, Sawamoto T, Zhang W et al. A phase 1, randomized ascending single-dose study of antagonist anti-human CD40 ASKP1240 in healthy subjects. Am J Transplant 2013;13(4):1040–1046. [DOI] [PubMed] [Google Scholar]
- 22.Imai A, Suzuki T, Sugitani A, Itoh T, Ueki S, Aoyagi T et al. A novel fully human anti-CD40 monoclonal antibody, 4D11, for kidney transplantation in cynomolgus monkeys. Transplantation 2007;84(8):1020–1028. [DOI] [PubMed] [Google Scholar]
- 23.Nashan BTH, van den Hoogen M, Berger S, Cibrik D, Mulgaonkar S, Leeser D, Alloway R, Patel A, Pratschke J, Sommerer C, Wiseman A, van Zuilen A, Laessing U, Rush J, Haraldsson B, Witzke O. CFZ533, a New Anti-CD40 mAB Demonstrates Comparable Efficacy and Better Renal Function versus Tacrolimus in De-Novo CNI-Free Kidney Transplantation. In. American Transplant Congress. 2018. [Google Scholar]
- 24.Ristov J, Espie P, Ulrich P, Sickert D, Flandre T, Dimitrova M et al. Characterization of the in vitro and in vivo properties of CFZ533, a blocking and non-depleting anti-CD40 monoclonal antibody. Am J Transplant 2018;18(12):2895–2904. [DOI] [PubMed] [Google Scholar]
- 25.Visvanathan S, Daniluk S, Ptaszynski R, Muller-Ladner U, Ramanujam M, Rosenstock B et al. Effects of BI 655064, an antagonistic anti-CD40 antibody, on clinical and biomarker variables in patients with active rheumatoid arthritis: a randomised, double-blind, placebo-controlled, phase IIa study. Ann Rheum Dis 2019;78(6):754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams AB, Shirasugi N, Jones TR, Durham MM, Strobert EA, Cowan S et al. Development of a chimeric anti-CD40 monoclonal antibody that synergizes with LEA29Y to prolong islet allograft survival. J Immunol 2005;174(1):542–550. [DOI] [PubMed] [Google Scholar]
- 27.Barr TA, Heath AW. Functional activity of CD40 antibodies correlates to the position of binding relative to CD154. Immunology 2001;102(1):39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankert KC, Oxley KL, Smith SM, Graham JP, de Boer M, Thewissen M et al. Induction of an altered CD40 signaling complex by an antagonistic human monoclonal antibody to CD40. J Immunol 2015;194(9):4319–4327. [DOI] [PubMed] [Google Scholar]
- 29.Yu X, Chan HTC, Orr CM, Dadas O, Booth SG, Dahal LN et al. Complex Interplay between Epitope Specificity and Isotype Dictates the Biological Activity of Anti-human CD40 Antibodies. Cancer Cell 2018;33(4):664–675 e664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pol E, Roos H, Markey F, Elwinger F, Shaw A, Karlsson R. Evaluation of calibration-free concentration analysis provided by Biacore systems. Anal Biochem 2016;510:88–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.