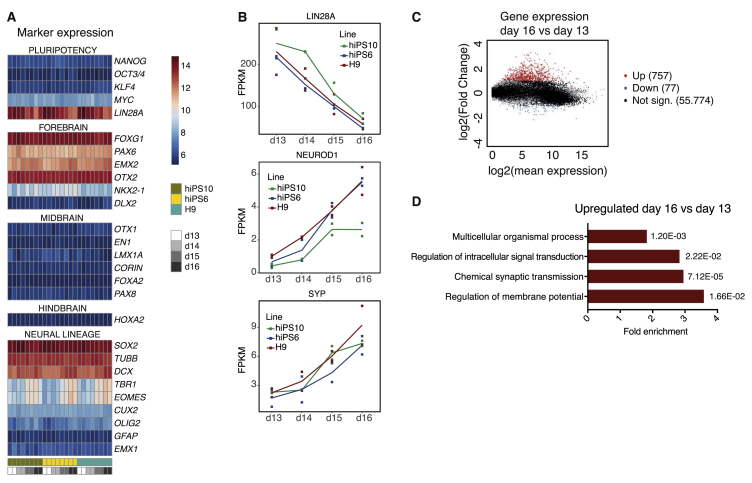
Figure 3.
Temporal transcriptome changes from day 13 to day 16. RNA-seq data of H9, hiPS6 and hiPS10 fbNPCs at day 13, 14, 15, and 16 of differentiation. (A) Heatmap displaying the marker expression profile between day 13 and day 16, two differentiation replicates per time-point. (B) Expression of LIN28A, NEUROD1, and SYP transcripts plotted as fragments per kilobase of transcript per million mapped reads (FPKM), the line represents average values for each time-point and the squares represent each differentiation replicate. (C) MA plot displaying significantly upregulated (p-adj. < 0.0001 & log2(FC) > 1) genes in day 16 compared to day 13 plotted in red, significantly downregulated (p-adj. < 0.0001 & log2(FC) < -1) genes in blue and non-significant genes in black. (D) Gene ontology analysis of upregulated genes (as shown in C) showing the fold enrichment and p-values for each parent term.