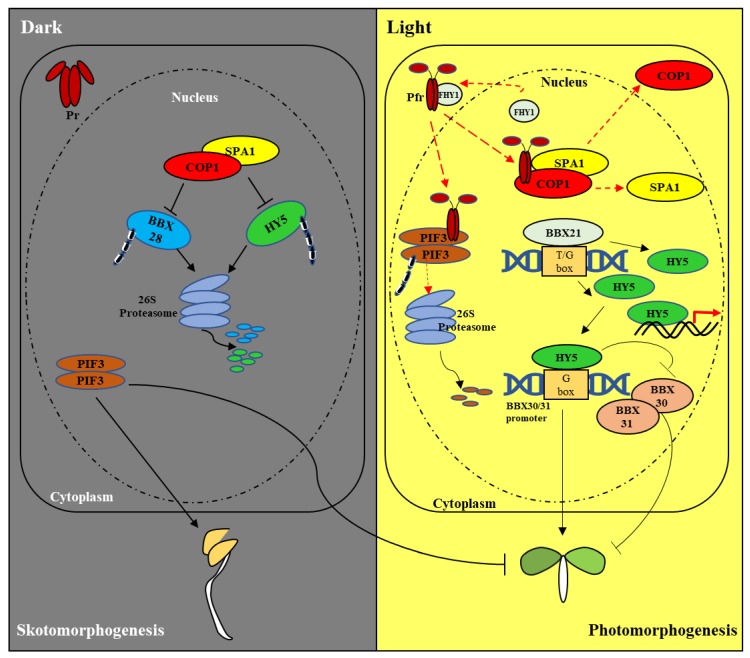
Figure 2.
A simplified view of the phytochrome-mediated light signaling pathway in A. thaliana. For simplicity, PIF3 (phytochrome interacting factor 3) was used as a representative of PIFs and SPA1 (suppressor of phyA-105 1) as a representative of the SPA proteins (SPA1 to SPA4). In the dark (left panel), phytochromes are synthesized as the inactive Pr form, remaining in the cytoplasm. Meanwhile, PIF proteins accumulate in the nucleus and negatively regulate the expression of genes involved in photomorphogenesis (shown as a T-headed line), allowing skotomorphogenesis (shown as an arrow-headed line). In addition, the COP1(constitutive photomorphogenesis protein 1)/SPA1 complex degrades HY5 (elongated hypocotyl 5) and BBX28 (B-box domain protein 28) via the ubiquitin/26S proteasome-mediated pathway to inhibit photomorphogenesis. Under light condition (right panel), photoactivated phytochromes (Pfr) accumulate in the nucleus (For phyA, FHY1 (far-red elongated hypocotyl 1) and FHL (FHY1-like) are the facilitators for the nuclear localization. Here, only FHY1 is shown for simplicity). In the nucleus, the Pfr form interacts with downstream signaling components such as PIF3 and the COP1/SPA1 complex. Phytochromes inactivate PIF3 via the 26S proteasome-mediated degradation pathway, and also inactivate the COP1/SPA1 complex by inducing dissociation, in which COP1 is subsequently exported to the cytoplasm. Among B-box proteins, BBX21 binds to the T/G box region of HY5 promoter and upregulates its expression. Thus, the inactivation of the COP1/SPA1 complex and BBX21 function contribute to the accumulation of HY5. In turn, HY5 induces the expression of light-responsive genes for photomorphogenesis (red arrow), and also suppresses BBX30 and BBX31, the negative regulators of photomorphogenesis, all of which promote photomorphogenic development.