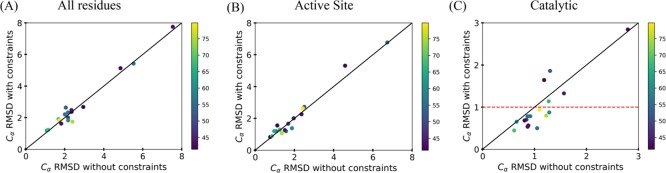
Figure 2.
Analysis of the Cα rmsd of the lowest five models for each enzyme in the benchmark with and without the incorporation of the CG constraints with a weight of 1. The rmsd was determined by comparing the target crystal structure to the models that were generated. Each point represents the average rmsd of the lowest five models, and the color of each point in the graphs represents the percent identity of the top template used for modeling. Any point seen below the line was seen as an improvement in modeling, on the line there was no change, and above the line, it was seen as a lack of improvement. (A) All residue rmsd. (B) Active site rmsd (residues within 8 Å of the ligand). (C) Catalytic residue rmsd.