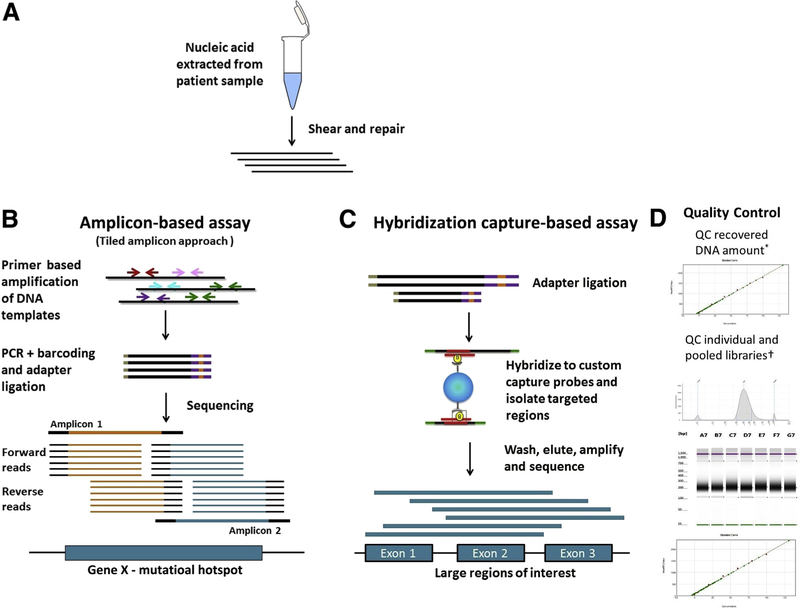
Figure 1.
High-level comparison of target enrichment workflow for amplicon and capture hybridization NGS assays. A: Nucleic acid is extracted and quantified. The DNA is sheared and repaired to generate fragments of uniform size distribution, and the fragment size can be monitored by gel electrophoresis or Agilent Bioanalyzer. B: Amplification-based assays: Target enrichment in amplification-based assays consists of PCR amplification of the desired region using primers. A tiled amplicon approach is depicted in which primers are designed to generate multiple overlapping amplicons of the same region to avoid allele dropout. The sequencing reads generated will have the same start and stop coordinates dictated by the primer design. C: Hybridization capture–based assays: Target enrichment in hybridization capture–based assays uses long biotinylated oligonucleotide probes complementary to a region of interest. Probes hybridize to target regions contained within larger fragments of DNA. As a result, regions flanking the target will also be isolated and sequenced. Targeted fragments are isolated using streptavidin magnetic beads, followed by washing, elution, amplification, and sequencing. The sequencing reads from these molecules will have unique start and stop coordinates when aligned to a reference, allowing identification and removal of PCR duplicates. D: Quality control: The size distribution pattern of the individual and pooled libraries are quality controlled using Agilent TapeStation and quantified using a Spectramax microplate reader. Example images as visualized using an Agilent Bioanalyzer and Spectramax microplate reader are shown. QC, quality control.