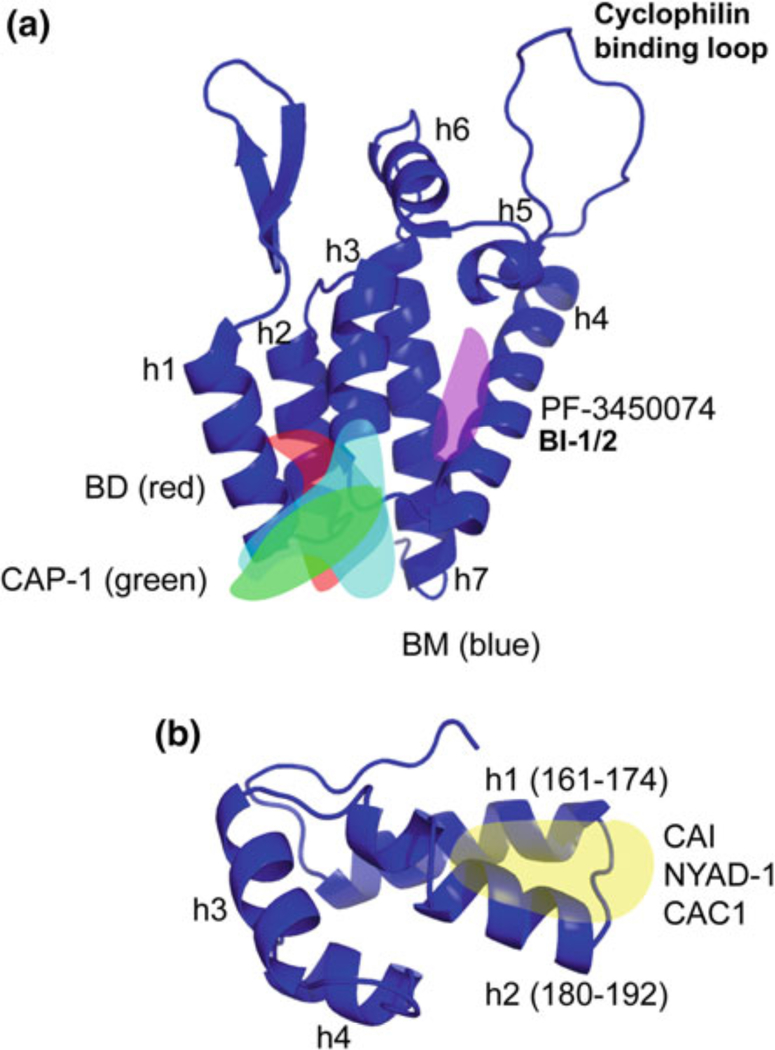
Fig. 3.
Crystal structures of the NTD (a) and CTD (b) of CA. Binding sites of CA inhibitors are indicated, where known, as transparent overlays. a The binding site of BM, BD, and CAP-1 is known as “site 1”; the binding site of PF-3450074 and BI-1/2 is known as “site 2.” b The binding site of CAI, NYAD-1, and CAC1 at the CTD dimer interface is indicated. Helix (h) numbers are indicated. Structures for NTD and CTD were generated in Pymol based on Protein Data Bank (PDB) coordinates 1GWP and 1A80, respectively (Gamble et al. 1997; Tang et al. 2002)