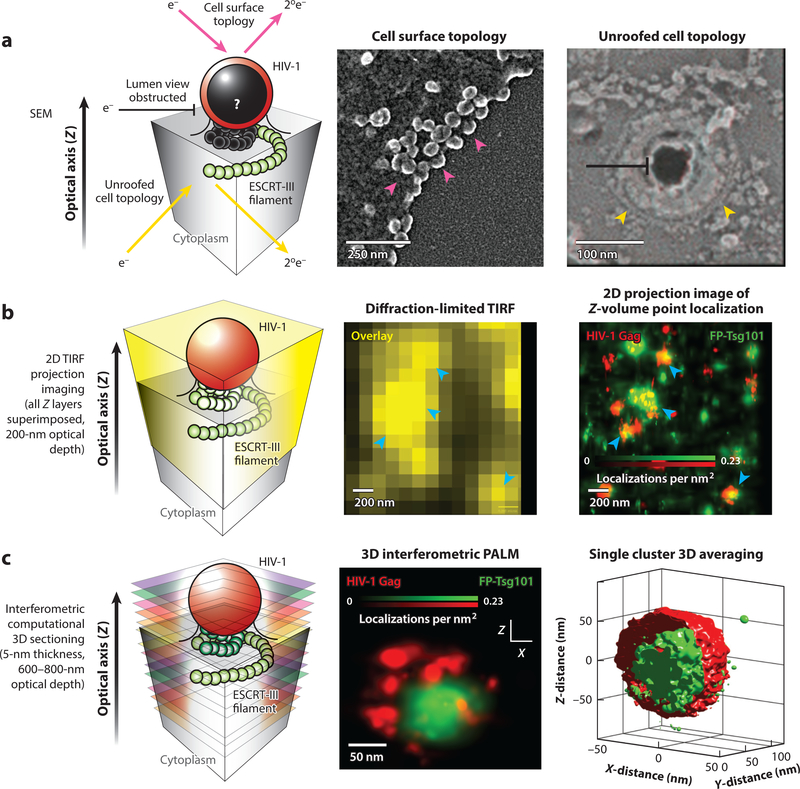
Figure 2.
Imaging modalities for interrogating the structural organization of the interface between HIV-1 Gag and the endosomal sorting complex required for transport (ESCRT) machinery. (a) Scanning electron microscopy (SEM) can interrogate exposed surface topologies of the HIV-1 budding process, probing either the surface of intact cells (magenta arrows; left micrograph) or the inner leaflet of the plasma membrane from mechanically unroofed cells (yellow arrows; right micrograph). This technique has very high spatial resolution (typically 1–5 nm) and easily resolves individual HIV-1 Gag particles (magenta arrowheads) and putative ESCRT machinery (yellow arrowheads). However, it does not enable direct visualization of the luminal contents of unperturbed HIV-1 Gag lattices as the budding membrane neck constricts (black). Right micrograph reproduced from Reference 45 with permission from Dr. Phyllis Hanson. (b) Total internal reflection fluorescence (TIRF) microscopy is an imaging technique that provides high contrast for structures of interest close to the plasma membrane (within ~200 nm). This technique, as with all fluorescence microscopy techniques, provides inherent molecular specificity due to direct fluorescent labeling of proteins of interest. TIRF microscopy is inherently diffraction limited and precludes resolution of objects much closer than 200–250 nm apart, resulting in the blurring of images between single HIV-1 assembly sites when the membrane is densely populated (blue arrowheads in left micrograph). 2D point localization microscopy techniques, such as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), bypass the diffraction limit and allow for nearly 10-fold improvements in resolution between HIV-1 assembly sites (blue arrowheads in right micrograph), as shown with HIV-1 Gag (red) and fluorescent protein (FP)-tagged Tsg101 (green; overlay yellow). TIRF and 2D point localization microscopy alone provide only projection images of all the fluorescent probes found within a ∼200-nm volume, which can complicate interpretation of 3D membrane remodeling processes such as HIV-1 budding. (c) Interferometric PALM (iPALM) utilizes light wave interference to extract the 3D position of a single fluorescent molecule, which enables computational sectioning of individual budding HIV-1 Gag lattices (red) and precise positioning of associated cellular components such as Tsg101 (green). As with all fluorescent techniques, any unlabeled structures, such as membranes, remain transparent, which enables luminal views into the HIV-1 Gag lattice. This technique also enables 3D single HIV-1 Gag cluster averaging of registered fluorescent channels for statistical assessment of ESCRT spatial distributions; at the right is an isosurfaced probability density image with n = 450 particles.