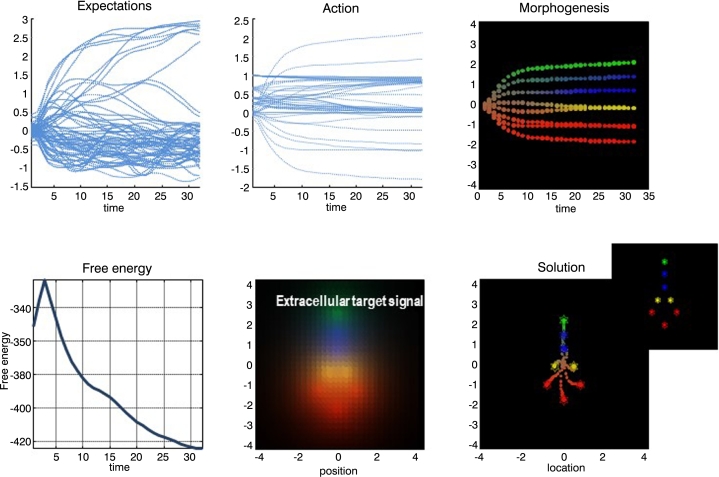
Fig. 4.
Self-assembly and active inference. This figure shows the results of a simulation of morphogenesis under active inference reported in [70]. This simulation used a gradient descent on variational free energy using a simple ensemble of eight cells; each of which had the same (pluripotential) generative model. This generative model predicted what each cell would sense and signal (chemotactically) for any given location in a ‘target morphology’ (lower middle panel – extracellular target signal; in other words, what each agent would expect to sense if it were at a particular location). By actively moving around, all the cells minimised their variational free energy (i.e., surprise) by inferring where they were, in relation to others. Because variational free energy is an extensive quantity, the free energy minimising arrangement of the ensemble is the target morphology. In other words, every cell has to ‘find its place’, at which point each cell minimises its own surprise about the signals it senses (because it knows its place) and the ensemble minimises the total free energy. The upper panels show the time courses of expectations about its place in the morphology (upper left), the associated active states mediating migration and signal expression (upper middle) and the resulting trajectories; projected onto the first (vertical) direction – and colour-coded to show differentiation (upper right). These trajectories progressively minimise total free energy (lower left panel). The lower right panel shows the ensuing configuration. Here, the trajectory is shown in small circles (for each time step). The insert corresponds to the target configuration. Please see [70] for further details.