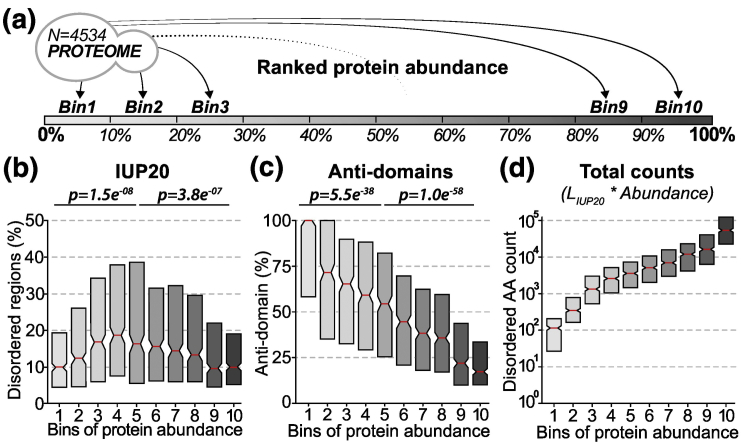
Fig. 1.
High-abundance proteins are depleted in disordered regions, yet they dominate the mass fraction of disordered residues in the cell. (a) The full yeast proteome, membrane proteins excluded, is split into 10 bins of protein abundance with equal sizes. Bins 1 and 10 correspond to the 10% proteins with lowest and highest abundance in S. cerevisiae. (b) Boxplot distribution of disorder content across the 10 bins of protein abundance. The disorder content (y-axis) corresponds to the percentage of disordered residues detected per protein using IUPred [84] (see Methods). P values indicate whether the median disorder content significantly differs between bin 5 against bin 1 or 10 (one-sided Wilcoxon signed-rank test). (c) Same as panel b, counting residues not covered by any Pfam [82], [85] or SUPERFAMILY [83], [86] structural domain (“anti-domain”). P values are calculated as in panel b. (d) Boxplot distribution of absolute amino acid counts in disordered regions with increasing levels of protein abundance. The absolute amino-acid counts (y-axis) correspond to the number of disordered amino acids multiplied by the cellular abundance of the protein (in ppm). Each box shows 50% of the density distribution where notches represent the 95% confidence interval around the median (red line).