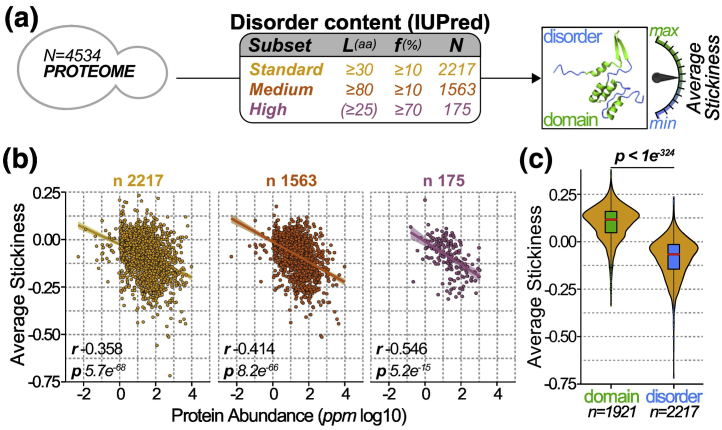
Fig. 2.
The stickiness of disordered regions is anti-correlated with protein abundance. (a) We defined three data sets of proteins with increasing disorder content, referred to as “standard,” “medium,” “high.” Proteins were included or excluded from each data set depending on their disorder content (L) and disorder fraction (f), as indicated in the panel table. The average stickiness of a protein was calculated by the mean score of all its amino acids in disordered regions. (b) Average protein stickiness (y-axis) as a function of protein abundance (x-axis) for disordered regions among the three data sets defined in panel a. The Spearman rank correlation coefficients (r) and the p values (p, Spearman’s rank correlation test) are indicated. The number of proteins (n) within each data set is given above each scatterplot. (c) Distribution of average protein stickiness (y-axis) calculated based on disordered regions (blue), or, for reference, calculated on domains (green) for the same proteins from the standard data set. Boxes correspond to 50% of the probability density around the median (red line). The p value indicates a significant difference between IDR-stickiness and domains stickiness (one-sided Wilcoxon signed-rank test).