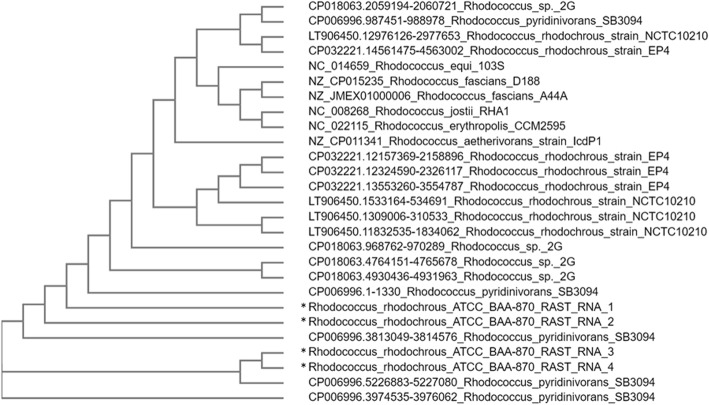
Fig. 1.
Phylogenetic tree created using rhodococcal 16S rRNA ClustalW sequence alignments. Neighbour joining, phylogenetic cladogram created using Phylogeny in ClustalW, and ClustalO multiple sequence alignment of R. rhodochrous ATCC BAA-870 16S rRNA genes and other closely matched genes from rhodococcal species. R. rhodochrous ATCC BAA-870 contains four copies of the 16S rRNA gene (labelled RNA_1 to RNA_4) and are indicated with an asterisk. For clarity, only closely matched BLAST results with greater than 95.5% sequence identity and those with complete 16S rRNA gene sequences, or from complete genomes, are considered. Additionally, 16S rRNA gene sequences (obtained from the NCBI gene database) from R. jostii RHA1, R. fascians A44A and D188, R. equi 103S, R. erythropolis CCM2595, and R. aetherivorans strain IcdP1 are included for comparison. Strain names are preceded by their NCBI accession number, as well as sequence position if there are multiple copies of the 16S rRNA gene in the same species