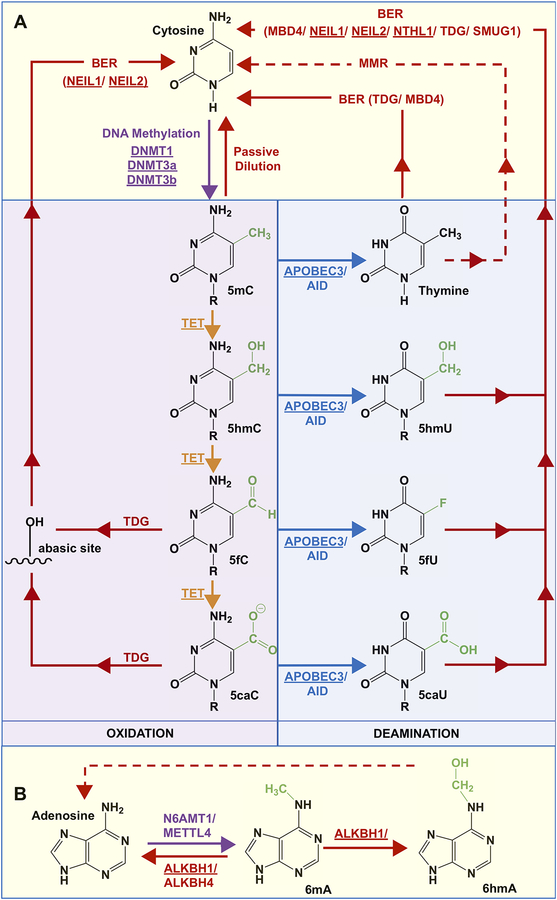
Figure 2:
Potential pathways of (de)methylation of cytosine and adenine. A) DNA methyltransferase (DNMT) enzymes 1, 3a, and 3b convert C to 5mC. Oxidation of 5mC by the TET enzymes results in 5hmC, 5fC, and 5caC; in the nucleus, the monofunctional TDG recognizes and catalyzes the removal of 5fC or 5caC, and the resulting abasic sites are converted to cytosine by NEIL-mediated BER. Other possibilities to restore the original cytosine base include active deamination of 5mC, 5hmC, 5fC and 5caC by AID or APOBEC3 to yield thymine, 5hmU, 5fU, and 5caU, respectively, which are excised by a DNA glycosylase and reverted to cytosine by BER. Given that several glycosylases have overlapping substrate preferences, MBD4, NEIL1, NEIL2, NTHL1, TDG, or SMUG1 can facilitate the repair of the deaminated bases. In the mitochondria given that TDG, SMUG1, and MBD4 are not present, 5mC that is deaminated to T yielding a T-G mispair can be repaired via mismatch repair, but this remains unknown (dotted red line). B) In the nucleus, A can be methylated to 6mA by N6AMT1/METTL4, and this process can be reversed by the ALKBH1/4 demethylases. N6-hydroxymethyladenine (6hmA) can also be formed by the oxidation of 6mA by ALKBH1, which may be converted to A via a mechanism that is yet unknown (dotted red line). Proteins that are underlined have been observed in the mitochondria.