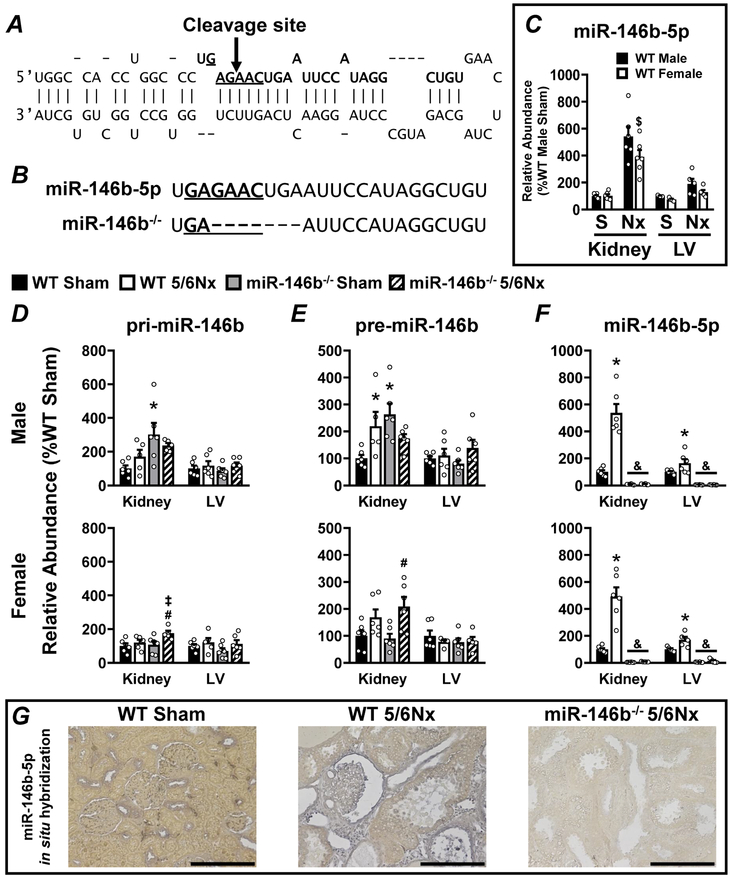
Figure 1: Efficient knockout of miR-146b-5p in SD rats in both the kidney and the LV in males and females.
CRISPR guide RNAs were designed to target within the seed sequence of Mir146b. (A) A diagram representing the stem-loop sequence of Mir146b highlights the seed sequence (underlined base pairs) of the mature miR-146b-5p sequence (base pairs in bold typeface). CRISPR-Cas9 targeting to this region resulted in a cleavage site within the seed sequence indicated by the arrow. (B) CRISPR-Cas9-mediated cleavage of Mir146b resulted in a 7-bp deletion overlapping the seed sequence of miR-146b-5p, indicated by dashed lines in bottom sequence. Mir146b gene products were characterized by qRT-PCR in both kidney and LV tissue: comparing (C) basal levels of miR-146b-5p in male vs. female WT, and the abundance of (D) primary Mir-146b transcript (pri-miR-146b), (E) the intermediate precursor (pre-miR-146b), and (F) the mature, bioactive, miR-146-5p in each experimental group. (G) ISH of miR-146b-5p in kidney tissue; representative images shown for WT sham, WT 5/6Nx and miR-146b−/− 5/6Nx. All data presented as mean ± SEM, n = 5-6/group; $ p<0.05 WT female vs. WT male; * p<0.05 WT 5/6Nx vs. WT Sham; # p<0.05 miR-146b−/− 5/6Nx vs. miR-146b−/− Sham; ‡ p<0.05 miR-146b−/− 5/6Nx vs. WT 5/6Nx; & p<0.05 miR-146b−/− vs. WT within all groups.