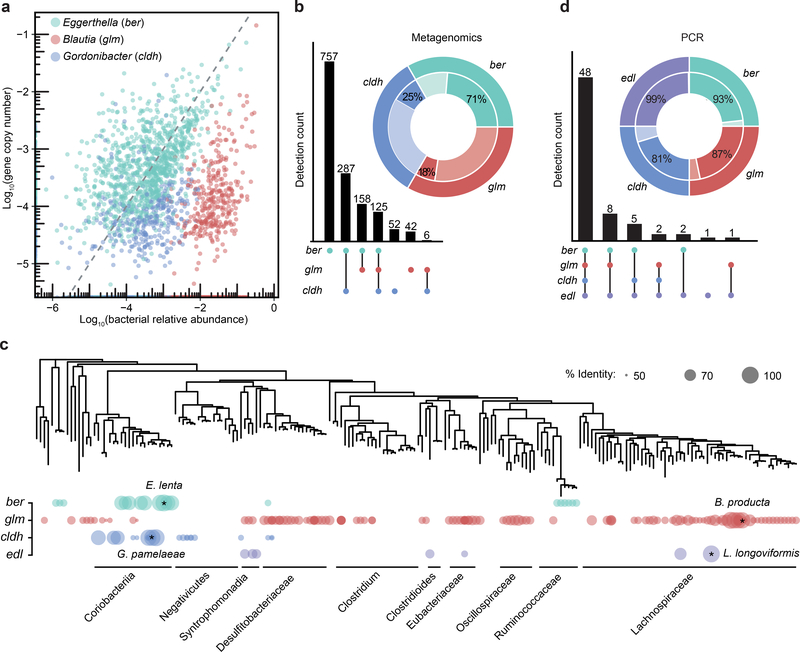
Figure 4. Genes linked to lignan metabolism are significantly correlated with bacterial host genera and prevalent in human gut microbiomes.
a, Correlation of putative lignan-metabolizing genes with host genera in diverse metagenomes (total subjects=1870). The dotted line (x=y) represents a perfect linear correlation. b, Co-occurrence of the lignan metabolizing pathway in metagenomes. The UpSet plot represents within-sample patterns in gene co-occurrence and the number of samples in which that pattern is detected (detection count). Inset: calculated prevalence. c, Phylogenetic distribution of proteins linked to lignan metabolism across gut microbial genomes. A reported hit is denoted by a colored circle with size proportional to the percentage identity to each query sequence. d, PCR detection of genes implicated in lignan metabolism in the stool samples of 68 adults living in Northern California (for one individual, no genes were identified; Supplementary Table 6).