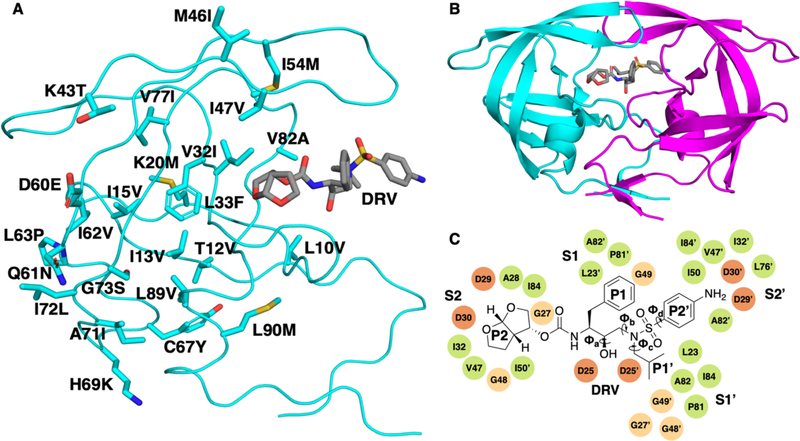
Figure 1.
(A) The 24 mutations of the KY variant (GenBank accession number: AY797430) mapped onto the structure of the HIV-1 protease monomer. (B) HIV-1 protease homodimer bound to DRV (represented in grey). Chain A is shown in cyan and chain B is shown in magenta. (C) 2D chemical structure of DRV with key dihedral angles labeled. These are the four dihedral angles that exhibited the greatest change between molecular dynamics simulations of the different KY variants. The residues corresponding to subsites S2 to S2′, where the different DRV moieties bind are labeled.