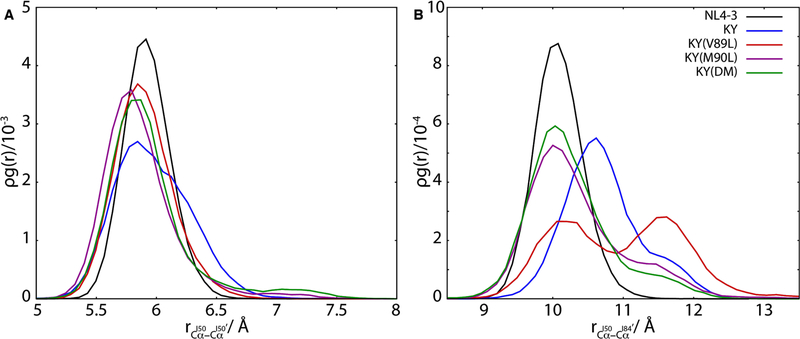
Figure 5.
Inter-residue distances near the catalytic site of HIV-1 protease are a gauge for inhibitor binding. The radial distribution function for the Cα atoms of residues I50 and I50′, shown in (A), exhibits an average shift to closer separations for the resistant variants relative to the wild-type NL4–3 protease. By contrast, the radial distribution function for the Cα atoms of residues I50 and I84′ (B) varies with resistance to DRV inhibition in these HIV-1 protease variants, with weaker binding variants visiting more open configurations.