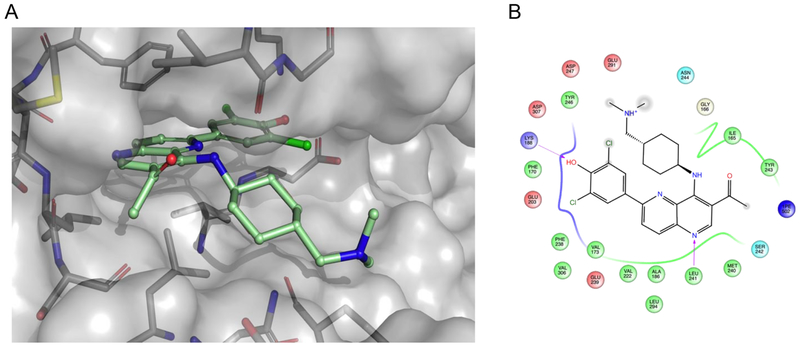
Figure 4.
Computational modeling of OTS167 5. (A) Modeled structure of OTS167 5 docked into the DYRK1A ATP-binding site. (B) Associated ligand interaction diagram between OTS167 5 and predicted key binding residues that are within 4 Å of the ligand. The gray spheres on atoms of OTS167 5 indicate regions that are solvent exposed. Arrows indicate key hydrogen bonds between the protein backbone (Leu241) and side-chain (Lys188) residues and the ligand. Negatively charged residues are indicated in red, positively charged residues are highlighted in blue, polar residues are highlighted in light blue, and hydrophobic residues are highlighted in green. Docking was performed using Schrodinger’s Glide software and PDB code 4MQ2 as the template for DYRK1A.55