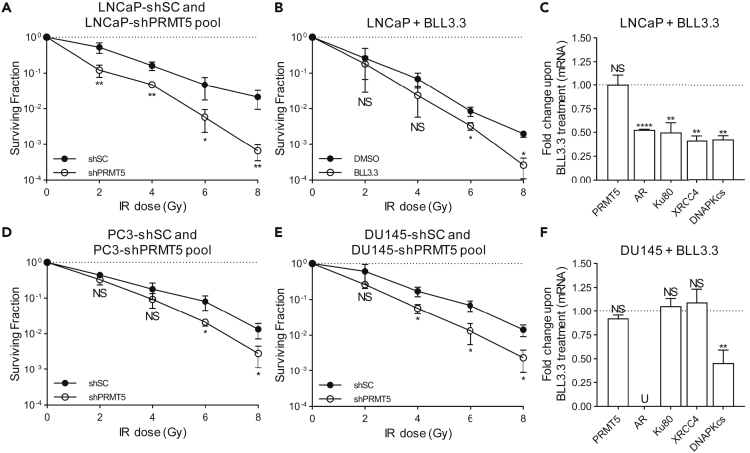
Figure 1.
Targeting PRMT5 Sensitizes Prostate Cancer Cells to IR in an AR-Independent Manner
(A, B, D, and E) Quantification of the surviving fraction via clonogenic assay immediately following the indicated dose of IR in the indicated cell lines. Dox treatment was used to express PRMT5-targeting shRNA (shPRMT5) or scramble control-targeting shRNA (shSC). BLL3.3 treatment was used to inhibit PRMT5 activity (A: LNCaP-shPRMT5/shSC, B: LNCaP + DMSO/BLL3.3, D: PC3-shPRMT5/shSC, E: DU145-shPRMT5/shSC).
(C and F) Quantification of mRNA via RT-qPCR 24 h post 2 Gy IR in LNCaP (C) and DU145 (F) cells. For each biological replicate, the value for BLL3.3 was normalized to the value for DMSO to calculate the fold change in mRNA expression upon PRMT5 inhibition.
Points in A, B, D, and E are the mean ± s.d. of three independent experiments. Bars in C and F are the mean ± s.d. of three independent experiments. Statistical analysis for A, B, D, and E comparing experimental with the control (“shSC” or “DMSO”) was performed using Welch's t test of log-transformed data, whereas statistical analysis for C and F comparing experimental with the control (“DMSO”) was performed using Welch's t test (*p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS p > 0.05, U = undetected).